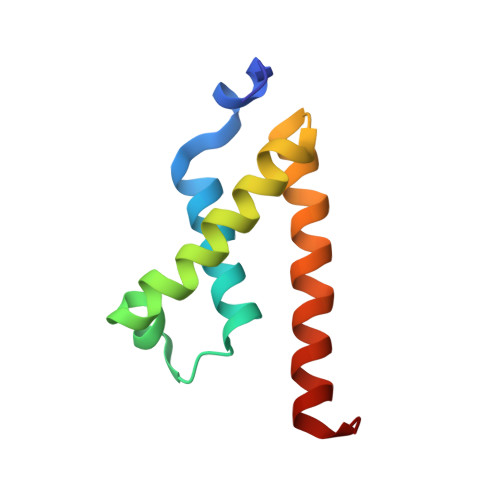
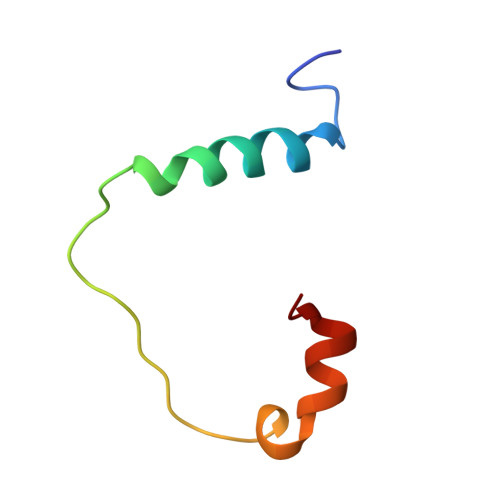
Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Wang, F., Marshall, C.B., Yamamoto, K., Li, G.Y., Gasmi-Seabrook, G.M., Okada, H., Mak, T.W., Ikura, M.(2012) Proc Natl Acad Sci U S A 109: 6078-6083
- PubMed: 22474372
- DOI: https://doi.org/10.1073/pnas.1119073109
- Primary Citation of Related Structures:
2LQH, 2LQI - PubMed Abstract:
Forkhead box class O 3a (FOXO3a) is a transcription factor and tumor suppressor linked to longevity that determines cell fate through activating transcription of cell differentiation, survival, and apoptotic genes. Recruitment of the coactivator CBP/p300 is a crucial step in transcription, and we revealed that in addition to conserved region 3 (CR3) of FOXO3a, the C-terminal segment of CR2 (CR2C) binds CBP/p300 and contributes to transcriptional activity. CR2C and CR3 of FOXO3a interact with the KIX domain of CBP/p300 at both "MLL" and "c-Myb" binding sites simultaneously. A FOXO3a CR2C-CR3 peptide in complex with KIX exists in equilibrium between two equally populated conformational states, one of which has CR2C bound to the MLL site and CR3 bound to the c-Myb site, whereas in the other, CR2C and CR3 bind the c-Myb and MLL sites, respectively. This promiscuous interaction between FOXO3a and CBP/p300 is further supported by additional binding sites on CBP/p300, namely, the TAZ1 and TAZ2 domains. In functional studies, our structure-guided mutagenesis showed that both CR2C and CR3 are involved in the activation of certain endogenous FOXO3a target genes. Further, phosphorylation of S626, a known AMP-dependent protein kinase target in CR3, increased affinity for CBP/p300 and the phosphomimetic mutation enhanced transactivation of luciferase. These findings underscore the significance of promiscuous multivalent interactions and posttranslational modification in the recruitment of transcriptional coactivators, which may allow transcription factors to adapt to various gene-specific genomic and chromatin structures and respond to cell signals.
Organizational Affiliation:
Ontario Cancer Institute, University Health Network, Toronto, ON, Canada M5G 1L7.