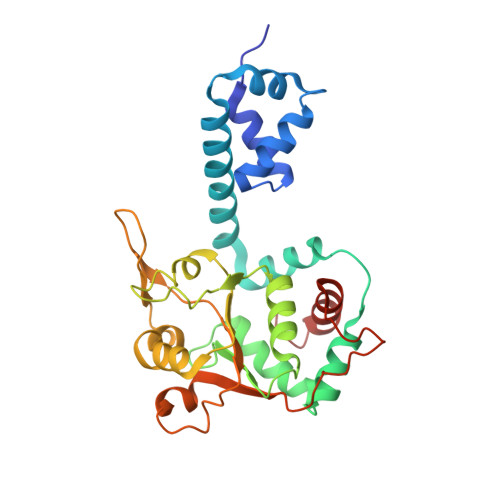
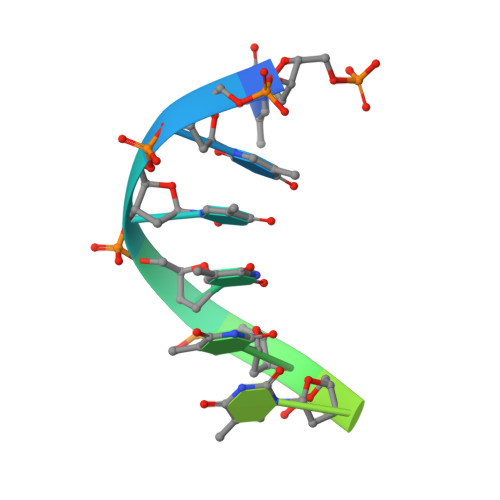
Mechanism of DNA translocation in a replicative hexameric helicase.
Enemark, E.J., Joshua-Tor, L.(2006) Nature 442: 270-275
- PubMed: 16855583
- DOI: https://doi.org/10.1038/nature04943
- Primary Citation of Related Structures:
2GXA - PubMed Abstract:
The E1 protein of papillomavirus is a hexameric ring helicase belonging to the AAA + family. The mechanism that couples the ATP cycle to DNA translocation has been unclear. Here we present the crystal structure of the E1 hexamer with single-stranded DNA discretely bound within the hexamer channel and nucleotides at the subunit interfaces. This structure demonstrates that only one strand of DNA passes through the hexamer channel and that the DNA-binding hairpins of each subunit form a spiral 'staircase' that sequentially tracks the oligonucleotide backbone. Consecutively grouped ATP, ADP and apo configurations correlate with the height of the hairpin, suggesting a straightforward DNA translocation mechanism. Each subunit sequentially progresses through ATP, ADP and apo states while the associated DNA-binding hairpin travels from the top staircase position to the bottom, escorting one nucleotide of single-stranded DNA through the channel. These events permute sequentially around the ring from one subunit to the next.
Organizational Affiliation:
W. M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, 1 Bungtown Road, Cold Spring Harbor, New York 11724, USA.