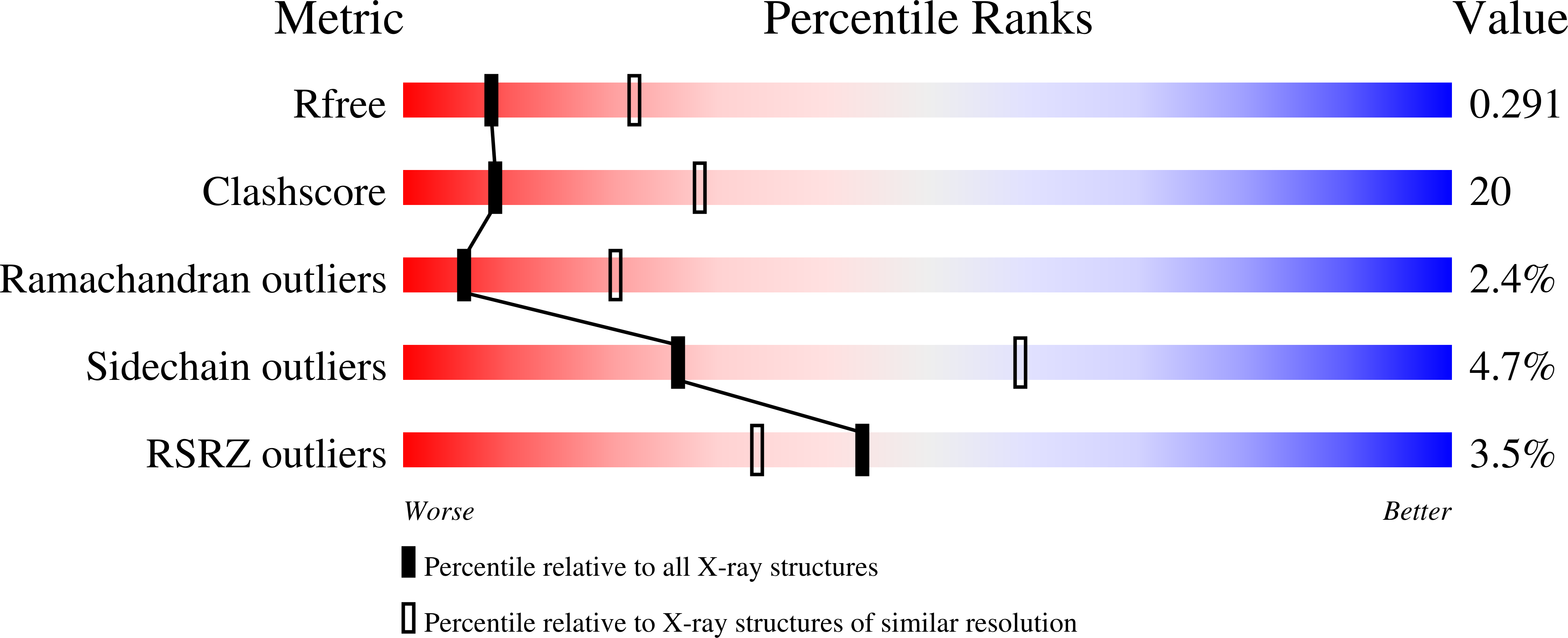
Structural basis of carbohydrate transfer activity by human UDP-GalNAc: polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10)
Kubota, T., Shiba, T., Sugioka, S., Furukawa, S., Sawaki, H., Kato, R., Wakatsuki, S., Narimatsu, H.(2006) J Mol Biol 359: 708-727
- PubMed: 16650853
- DOI: https://doi.org/10.1016/j.jmb.2006.03.061
- Primary Citation of Related Structures:
2D7I, 2D7R - PubMed Abstract:
Mucin-type O-glycans are important carbohydrate chains involved in differentiation and malignant transformation. Biosynthesis of the O-glycan is initiated by the transfer of N-acetylgalactosamine (GalNAc) which is catalyzed by UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferases (pp-GalNAc-Ts). Here we present crystal structures of the pp-GalNAc-T10 isozyme, which has specificity for glycosylated peptides, in complex with the hydrolyzed donor substrate UDP-GalNAc and in complex with GalNAc-serine. A structural comparison with uncomplexed pp-GalNAc-T1 suggests that substantial conformational changes occur in two loops near the catalytic center upon donor substrate binding, and that a distinct interdomain arrangement between the catalytic and lectin domains forms a narrow cleft for acceptor substrates. The distance between the catalytic center and the carbohydrate-binding site on the lectin beta sub-domain influences the position of GalNAc glycosylation on GalNAc-glycosylated peptide substrates. A chimeric enzyme in which the two domains of pp-GalNAc-T10 are connected by a linker from pp-GalNAc-T1 acquires activity toward non-glycosylated acceptors, identifying a potential mechanism for generating the various acceptor specificities in different isozymes to produce a wide range of O-glycans.
Organizational Affiliation:
Glycogene Function Team of Research Center for Glycoscience (RCG), National Institute of Advanced Industrial Science and Technology (AIST), Central 2, 1-1-1 Umezono, Tsukuba, Ibaraki 305-8568, Japan.