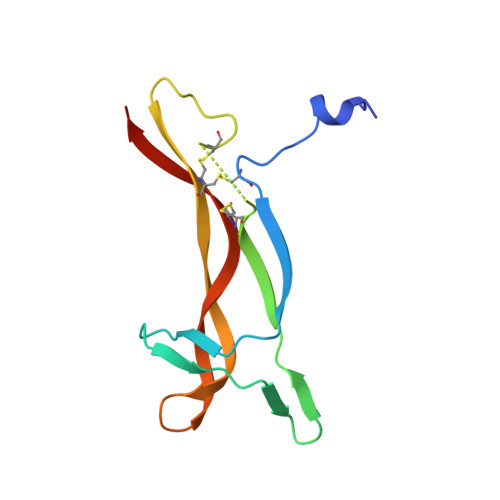
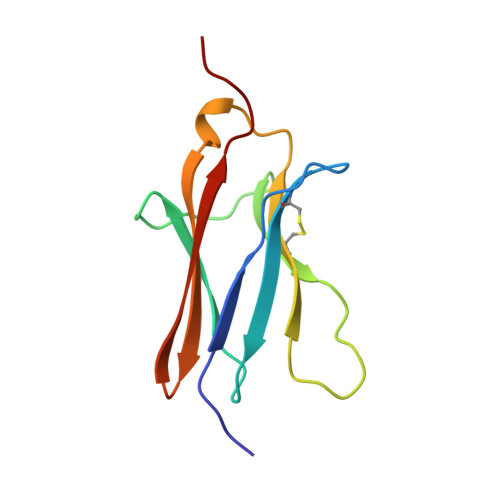
Crystal structure of nerve growth factor in complex with the ligand-binding domain of the TrkA receptor.
Wiesmann, C., Ultsch, M.H., Bass, S.H., de Vos, A.M.(1999) Nature 401: 184-188
- PubMed: 10490030
- DOI: https://doi.org/10.1038/43705
- Primary Citation of Related Structures:
1WWW - PubMed Abstract:
Nerve growth factor (NGF) is involved in a variety of processes involving signalling, such as cell differentiation and survival, growth cessation and apoptosis of neurons. These events are mediated by NGF as a result of binding to its two cell-surface receptors, TrkA and p75. TrkA is a receptor with tyrosine kinase activity that forms a high-affinity binding site for NGF. Of the five domains comprising its extracellular portion, the immunoglobulin-like domain proximal to the membrane (TrkA-d5 domain) is necessary and sufficient for NGF binding. Here we present the crystal structure of human NGF in complex with human TrkA-d5 at 2.2 A resolution. The ligand-receptor interface consists of two patches of similar size. One patch involves the central beta-sheet that forms the core of the homodimeric NGF molecule and the loops at the carboxy-terminal pole of TrkA-d5. The second patch comprises the amino-terminal residues of NGF, which adopt a helical conformation upon complex formation, packing against the 'ABED' sheet of TrkA-d5. The structure is consistent with results from mutagenesis experiments for all neurotrophins, and indicates that the first patch may constitute a conserved binding motif for all family members, whereas the second patch is specific for the interaction between NGF and TrkA.
Organizational Affiliation:
Department of Protein Engineering, Genentech, Inc., South San Francisco, California 94080, USA.