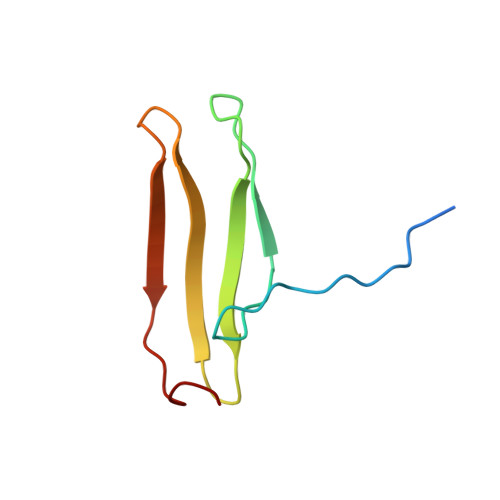
Solution structure of an Arabidopsis WRKY DNA binding domain.
Yamasaki, K., Kigawa, T., Inoue, M., Tateno, M., Yamasaki, T., Yabuki, T., Aoki, M., Seki, E., Matsuda, T., Tomo, Y., Hayami, N., Terada, T., Shirouzu, M., Tanaka, A., Seki, M., Shinozaki, K., Yokoyama, S.(2005) Plant Cell 17: 944-956
- PubMed: 15705956
- DOI: https://doi.org/10.1105/tpc.104.026435
- Primary Citation of Related Structures:
1WJ2 - PubMed Abstract:
The WRKY proteins comprise a major family of transcription factors that are essential in pathogen and salicylic acid responses of higher plants as well as a variety of plant-specific reactions. They share a DNA binding domain, designated as the WRKY domain, which contains an invariant WRKYGQK sequence and a CX4-5CX22-23HXH zinc binding motif. Herein, we report the NMR solution structure of the C-terminal WRKY domain of the Arabidopsis thaliana WRKY4 protein. The structure consists of a four-stranded beta-sheet, with a zinc binding pocket formed by the conserved Cys/His residues located at one end of the beta-sheet, revealing a novel zinc and DNA binding structure. The WRKYGQK residues correspond to the most N-terminal beta-strand, kinked in the middle of the sequence by the Gly residue, which enables extensive hydrophobic interactions involving the Trp residue and contributes to the structural stability of the beta-sheet. Based on a profile of NMR chemical shift perturbations, we propose that the same strand enters the DNA groove and forms contacts with the DNA bases.
Organizational Affiliation:
Age Dimension Research Center, National Institute of Advanced Industrial Science and Technology, Tsukuba 305-8566, Japan. k-yamasaki@aist.go.jp