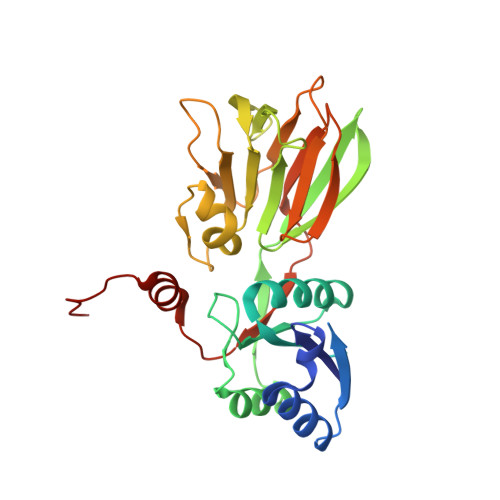
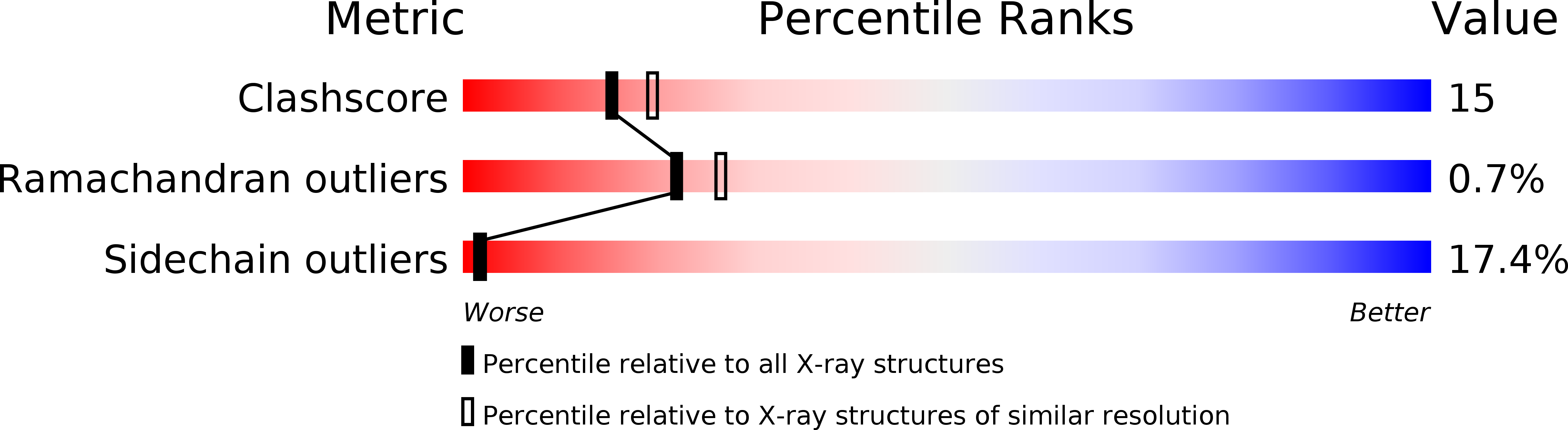A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis
Garavaglia, S., Raffaelli, N., Finaurini, L., Magni, G., Rizzi, M.(2004) J Biol Chem 279: 40980-40986
- PubMed: 15269221
- DOI: https://doi.org/10.1074/jbc.M406586200
- Primary Citation of Related Structures:
1U0R, 1U0T - PubMed Abstract:
NAD kinase catalyzes the magnesium-dependent phosphorylation of NAD, representing the sole source of freshly synthesized NADP in all organisms. The enzyme is essential for the growth of the deadly multidrug-resistant pathogen Mycobacterium tuberculosis and is an attractive target for novel antitubercular agents. The crystal structure of NAD kinase has been solved by multiwavelength anomalous dispersion at a resolution of 2.3 A in its T state. Two crystal forms have been obtained revealing either a dimer or a tetramer. The enzyme architecture discloses a novel molecular arrangement, with each subunit consisting of an alpha/beta N-terminal domain and a C-terminal 12-stranded beta sandwich domain, connected by swapped beta strands. The C-terminal domain shows a striking internal approximate 222 symmetry and an unprecedented topology, revealing a novel fold within the family of all beta structures. The catalytic site is located in the long crevice that defines the interface between the domains. The conserved GGDG structural fingerprint of the catalytic site is reminiscent of the related region in 6-phosphofructokinase, supporting the hypothesis that NAD kinase belongs to a newly reported superfamily of kinases.
Organizational Affiliation:
Dipartimento di Scienze Chimiche, Alimentari, Farmaceutiche, Farmacologiche-Istituto Nazionale Fisica della Materia, University of Piemonte Orientale "Amedeo Avogadro," Via Bovio 6, 28100 Novara, Italy.