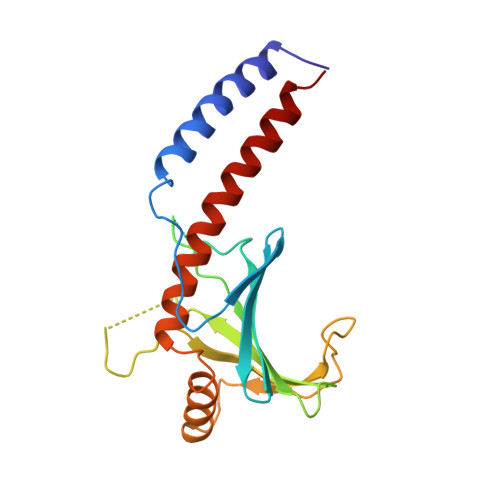
Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein.
Jiang, J., Zhang, Y., Krainer, A.R., Xu, R.M.(1999) Proc Natl Acad Sci U S A 96: 3572-3577
- PubMed: 10097078
- DOI: https://doi.org/10.1073/pnas.96.7.3572
- Primary Citation of Related Structures:
1P32 - PubMed Abstract:
Human p32 (also known as SF2-associated p32, p32/TAP, and gC1qR) is a conserved eukaryotic protein that localizes predominantly in the mitochondrial matrix. It is thought to be involved in mitochondrial oxidative phosphorylation and in nucleus-mitochondrion interactions. We report the crystal structure of p32 determined at 2.25 A resolution. The structure reveals that p32 adopts a novel fold with seven consecutive antiparallel beta-strands flanked by one N-terminal and two C-terminal alpha-helices. Three monomers form a doughnut-shaped quaternary structure with an unusually asymmetric charge distribution on the surface. The implications of the structure on previously proposed functions of p32 are discussed and new specific functional properties are suggested.
Organizational Affiliation:
W. M. Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.