CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs.
Kitadokoro, K., Bordo, D., Galli, G., Petracca, R., Falugi, F., Abrignani, S., Grandi, G., Bolognesi, M.(2001) EMBO J 20: 12-18
- PubMed: 11226150
- DOI: https://doi.org/10.1093/emboj/20.1.12
- Primary Citation of Related Structures:
1G8Q - PubMed Abstract:
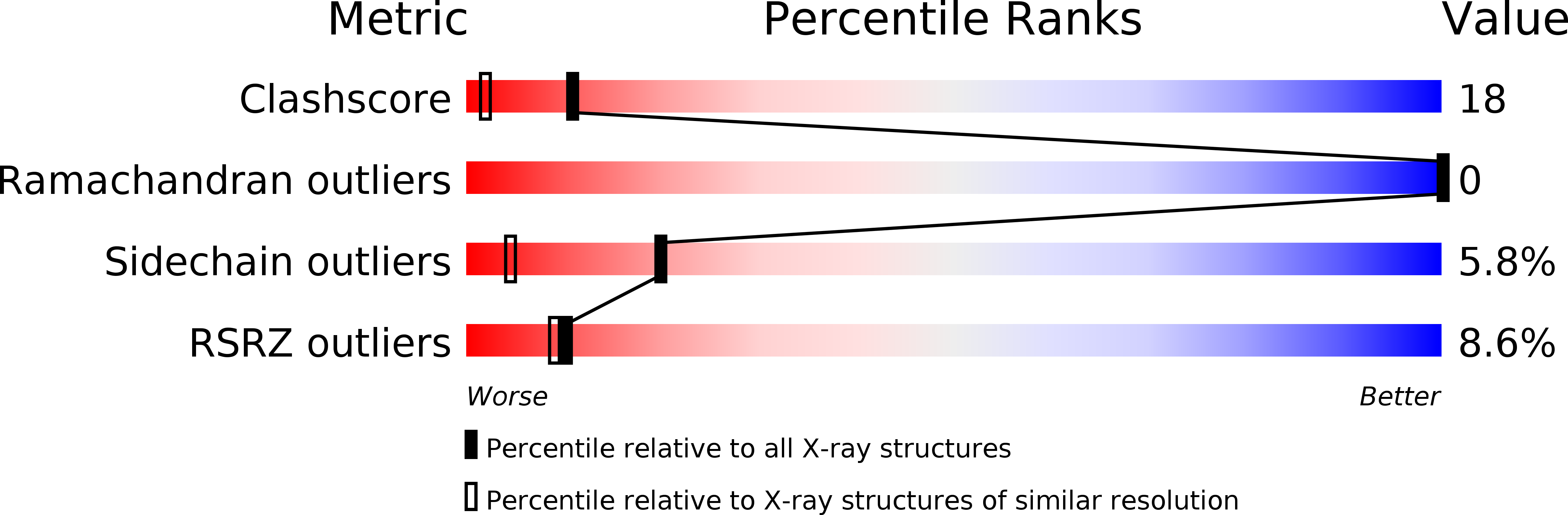
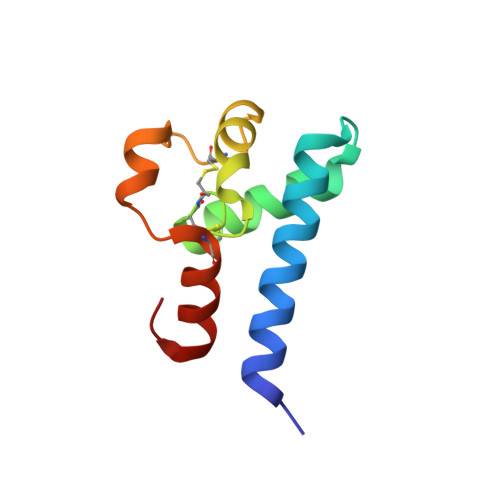
Human CD81, a known receptor for hepatitis C virus envelope E2 glycoprotein, is a transmembrane protein belonging to the tetraspanin family. The crystal structure of human CD81 large extracellular domain is reported here at 1.6 A resolution. Each subunit within the homodimeric protein displays a mushroom-like structure, composed of five alpha-helices arranged in 'stalk' and 'head' subdomains. Residues known to be involved in virus binding can be mapped onto the head subdomain, providing a basis for the design of antiviral drugs and vaccines. Sequence analysis of 160 tetraspanins indicates that key structural features and the new protein fold observed in the CD81 large extracellular domain are conserved within the family. On these bases, it is proposed that tetraspanins may assemble at the cell surface into homo- and/or hetero-dimers through a conserved hydrophobic interface located in the stalk subdomain, while interacting with other liganding proteins, including hepatitis C virus E2, through the head subdomain. The topology of such interactions provides a rationale for the assembly of the so-called tetraspan-web.
Organizational Affiliation:
Department of Physics, INFM and Advanced Biotechnology Center, University of Genoa, Via Dodecaneso, 33, I-16146 Genova, Switzerland.