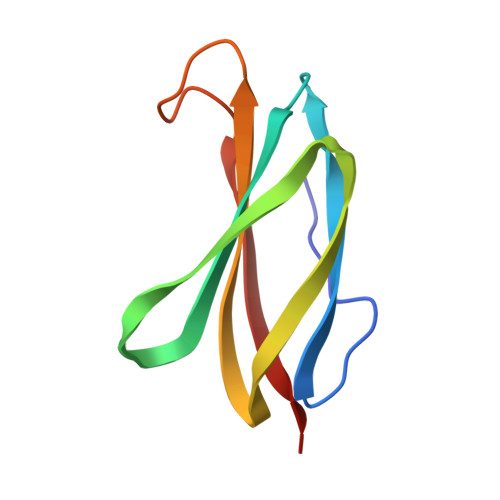
Crystal structure of the tenth type III cell adhesion module of human fibronectin.
Dickinson, C.D., Veerapandian, B., Dai, X.P., Hamlin, R.C., Xuong, N.H., Ruoslahti, E., Ely, K.R.(1994) J Mol Biol 236: 1079-1092
- PubMed: 8120888
- DOI: https://doi.org/10.1016/0022-2836(94)90013-2
- Primary Citation of Related Structures:
1FNA - PubMed Abstract:
The crystal structure of the cell adhesion module of fibronectin (FNIII10) has been determined at 1.8 A resolution. A recombinant fragment corresponding to the tenth type III module of human fibronectin was crystallized in space group P2(1) with a = 30.7, b = 35.1 and c = 37.7 A and beta = 107 degrees. The structure was determined by molecular replacement and refined by least squares methods. The crystallographic R-factor for the final model of the 91 amino acid module plus 56 solvent atoms is 0.18 for 10 to 1.8 A data. The module consists of two layers of beta-sheet, one with three antiparallel strands and the other with four antiparallel strands. The beta-sheets enclose a hydrophobic core of 24 amino acid side-chains. The module contains the RGD cell recognition sequence in a flexible loop connecting two beta-strands. The tertiary structure of the FNIII10 module has been used to develop a structure-based sequence alignment of 17 type III modules in fibronectin based on the striking conservation of homologous hydrophobic residues. A similar pattern of homologous alternating hydrophobic residues is also evident in a comparison of type III modules in proteins unrelated to fibronectin such as cytokine receptors and muscle proteins.
Organizational Affiliation:
Cancer Research Center, La Jolla Cancer Research Foundation, CA 92037.