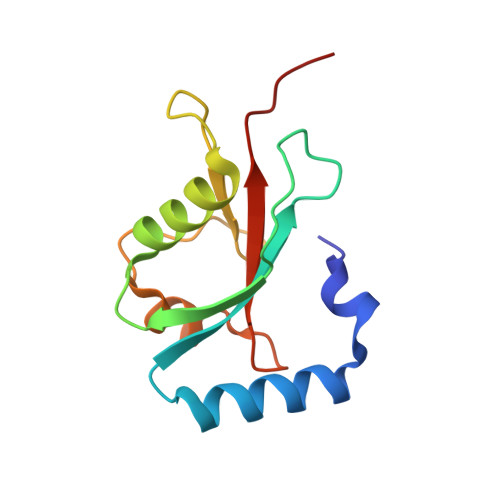
Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p.
Paz, Y., Elazar, Z., Fass, D.(2000) J Biol Chem 275: 25445-25450
- PubMed: 10856287
- DOI: https://doi.org/10.1074/jbc.C000307200
- Primary Citation of Related Structures:
1EO6 - PubMed Abstract:
The GATE-16 protein participates in intra-Golgi transport and can associate with the N-ethylmaleimide-sensitive fusion protein and with Golgi SNAREs. The yeast ortholog of GATE-16 is the autophagocytosis factor Aut7p. GATE-16 is also closely related to the GABA receptor-associated protein (GABARAP), which has been proposed to cluster neurotransmitter receptors by mediating interaction with the cytoskeleton, and to the light chain-3 subunit of the neuronal microtubule-associated protein complex. Here, we present the crystal structure of GATE-16 refined to 1.8 A resolution. GATE-16 contains a ubiquitin fold decorated by two additional N-terminal helices. Proteins with strong structural similarity but no detectable sequence homology to GATE-16 include Ras effectors that mediate diverse downstream functions, but each interacts with Ras by forming pseudo-continuous beta-sheets. The GATE-16 surface suggests that it binds its targets in a similar manner. Moreover, a second potential protein-protein interaction site on GATE-16 may explain the adapter activity observed for members of the GATE-16 family.
Organizational Affiliation:
Department of Structural Biology and Biological Chemistry, Weizmann Institute of Science, Rehovot, Israel.