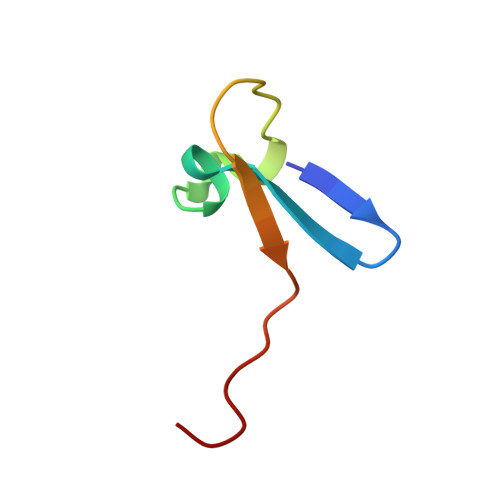
Solution structure of a zinc domain conserved in yeast copper-regulated transcription factors.
Turner, R.B., Smith, D.L., Zawrotny, M.E., Summers, M.F., Posewitz, M.C., Winge, D.R.(1998) Nat Struct Biol 5: 551-555
- PubMed: 9665167
- DOI: https://doi.org/10.1038/805
- Primary Citation of Related Structures:
1CO4 - PubMed Abstract:
The three dimensional structure of the N-terminal domain (residues 1-42) of the copper-responsive transcription factor Amtl from Candida glabrata has been determined by two-dimensional 1H-correlated nuclear magnetic resonance (NMR) methods. The domain contains an array of zinc-binding residues (Cys-X2-Cys-X8-Cys-X-His) that is conserved among a family of Cu-responsive transcription factors. The structure is unlike those of previously characterized zinc finger motifs, and consists of a three-stranded antiparallel beta-sheet with two short helical segments that project from one end of the beta-sheet. Conserved residues at positions 16, 18 and 19 form a basic patch that may be important for DNA binding.
Organizational Affiliation:
Howard HUghes Medical Institute, University of Maryland Baltimore County 21250, USA.