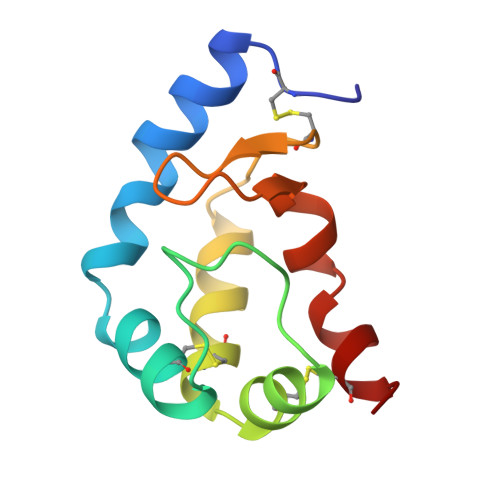
The 2.1 A structure of an elicitin-ergosterol complex: a recent addition to the Sterol Carrier Protein family.
Boissy, G., O'Donohue, M., Gaudemer, O., Perez, V., Pernollet, J.C., Brunie, S.(1999) Protein Sci 8: 1191-1199
- PubMed: 10386869
- DOI: https://doi.org/10.1110/ps.8.6.1191
- Primary Citation of Related Structures:
1BXM - PubMed Abstract:
Elicitins, produced by most of the phytopathogenic fungi of the genus Phytophthora, provoke in tobacco both remote leaf necrosis and the induction of a resistance against subsequent attack by various microorganisms. Despite the recent description of the three-dimensional crystal structure of cryptogein (CRY), the molecular basis of the interactions between Phytophthora and plants largely remains unknown. The X-ray crystal structure, refined at 2.1 A, of a ligand complexed, mutated CRY, K13H, is reported. Analysis of this structure reveals that CRY is able to encapsulate a ligand that induces only a minor conformational change in the protein structure. The ligand has been identified as an ergosterol by gas chromatographic analysis coupled with mass spectrometry analysis. This result is consistent with biochemical data that have shown that elicitins are a distinct class of Sterol Carrier Proteins (SCP). Data presented here provide the first structural description of the pertinent features of the elicitin sterol interaction and permit a reassessment of the importance of both the key residue 13 and the mobility of the omega loop for the accessibility of the sterol to the cavity. The biological implications thereof are discussed. This paper reports the first structure of a SCP/sterol complex.
Organizational Affiliation:
Unité de Recherche Biochimie & Structure des Protéines, INRA, Jouy-en-Josas, France.