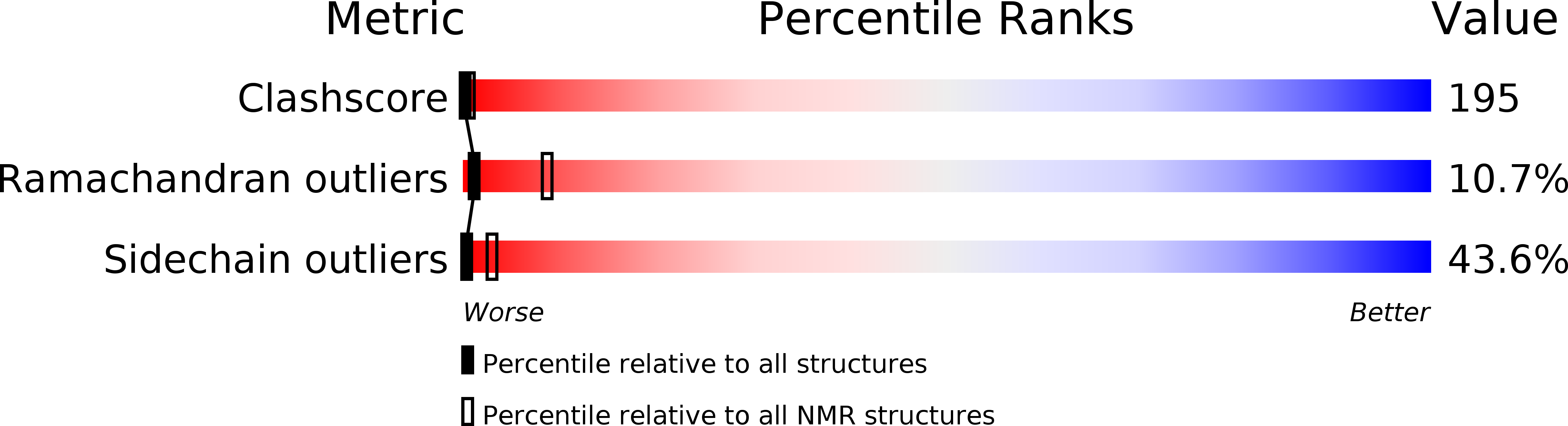1ACZ
GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN COMPLEX WITH CYCLODEXTRIN, NMR, 5 STRUCTURES
- PDB DOI: https://doi.org/10.2210/pdb1ACZ/pdb
- Classification: POLYSACCHARIDE DEGRADATION
- Organism(s): Aspergillus niger
- Expression System: Aspergillus niger
- Mutation(s): No
- Deposited: 1997-02-10 Released: 1997-07-07
Experimental Data Snapshot
- Method: SOLUTION NMR
- Conformers Calculated: 100
- Conformers Submitted: 5
- Selection Criteria: RANDOM FROM 81 GOOD STRUCTURES
wwPDB Validation 3D Report Full Report
This is version 2.0 of the entry. See complete history.
Macromolecules
Find similar proteins by:
(by identity cutoff) | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GLUCOAMYLASE | 108 | Aspergillus niger | Mutation(s): 0 EC: 3.2.1.3 |  | |
UniProt | |||||
Find proteins for P69328 (Aspergillus niger) Explore P69328 Go to UniProtKB: P69328 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P69328 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Oligosaccharides
Biologically Interesting Molecules (External Reference) 1 Unique
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_900012 Query on PRD_900012 | B, C | beta-cyclodextrin | Oligosaccharide / Drug delivery |  | |
Experimental Data & Validation
Experimental Data
- Method: SOLUTION NMR
- Conformers Calculated: 100
- Conformers Submitted: 5
- Selection Criteria: RANDOM FROM 81 GOOD STRUCTURES
Entry History
Deposition Data
- Released Date: 1997-07-07 Deposition Author(s): Sorimachi, K., Le Gal-Coeffet, M.-F., Williamson, G., Archer, D.B., Williamson, M.P.
Revision History (Full details and data files)
- Version 1.0: 1997-07-07
Type: Initial release - Version 1.1: 2008-03-24
Changes: Version format compliance - Version 1.2: 2011-07-13
Changes: Version format compliance - Version 2.0: 2020-07-29
Type: Remediation
Reason: Carbohydrate remediation
Changes: Atomic model, Data collection, Derived calculations, Other, Structure summary