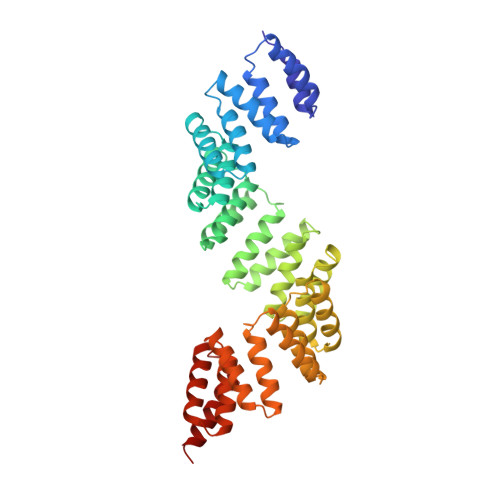
The Superhelical Tpr-Repeat Domain of O-Linked Glcnac Transferase Exhibits Structural Similarities to Importin Alpha
Jinek, M., Rehwinkel, J., Lazarus, B.D., Izaurralde, E., Hanover, J.A., Conti, E.(2004) Nat Struct Mol Biol 11: 1001
- PubMed: 15361863
- DOI: https://doi.org/10.1038/nsmb833
- Primary Citation of Related Structures:
1W3B - PubMed Abstract:
Addition of N-acetylglucosamine (GlcNAc) is a ubiquitous form of intracellular glycosylation catalyzed by the conserved O-linked GlcNAc transferase (OGT). OGT contains an N-terminal domain of tetratricopeptide (TPR) repeats that mediates the recognition of a broad range of target proteins. Components of the nuclear pore complex are major OGT targets, as OGT depletion by RNA interference (RNAi) results in the loss of GlcNAc modification at the nuclear envelope. To gain insight into the mechanism of target recognition, we solved the crystal structure of the homodimeric TPR domain of human OGT, which contains 11.5 TPR repeats. The repeats form an elongated superhelix. The concave surface of the superhelix is lined by absolutely conserved asparagines, in a manner reminiscent of the peptide-binding site of importin alpha. Based on this structural similarity, we propose that OGT uses an analogous molecular mechanism to recognize its targets.
Organizational Affiliation:
Structural and Computational Biology, European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.