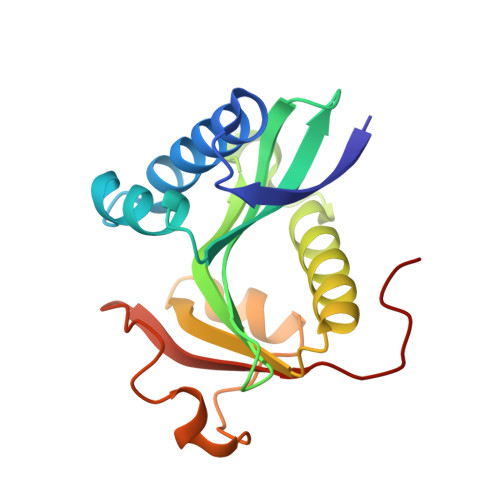
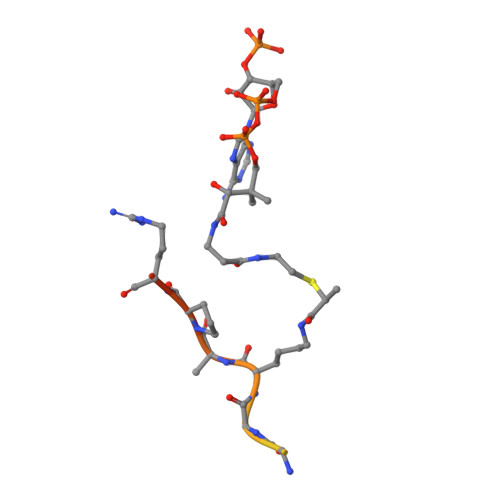
Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor.
Poux, A.N., Cebrat, M., Kim, C.M., Cole, P.A., Marmorstein, R.(2002) Proc Natl Acad Sci U S A 99: 14065-14070
- PubMed: 12391296
- DOI: https://doi.org/10.1073/pnas.222373899
- Primary Citation of Related Structures:
1M1D - PubMed Abstract:
Histone acetyltransferases (HATs) use acetyl CoA to acetylate target lysine residues within histones and other transcription factors, such as the p53 tumor suppressor, to promote gene activation. HAT enzymes fall into subfamilies with divergence in sequence and substrate preference. Several HAT proteins have been implicated in human cancer. We have previously reported on the preparation of peptide-CoA conjugate inhibitors with distinct specificities for the p300/CBP [cAMP response element binding protein (CREB)-binding protein] or GCN5 HAT subfamilies. Here we report on the crystal structure of the GCN5 HAT bound to a peptide-CoA conjugate containing CoA covalently attached through an isopropionyl linker to Lys-14 of a 20-aa N-terminal fragment of histone H3. Surprisingly, the structure reveals that the H3 portion of the inhibitor is bound outside of the binding site for the histone substrate and that only five of the 20 aa residues of the inhibitor are ordered. Rearrangements within the C-terminal region of the GCN5 protein appear to mediate this peptide displacement. Mutational and enzymatic data support the hypothesis that the observed structure corresponds to a late catalytic intermediate. The structure also provides a structural scaffold for the design of HAT-specific inhibitors that may have therapeutic applications for the treatment of HAT-mediated cancers.
Organizational Affiliation:
The Wistar Institute, and Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104, USA.