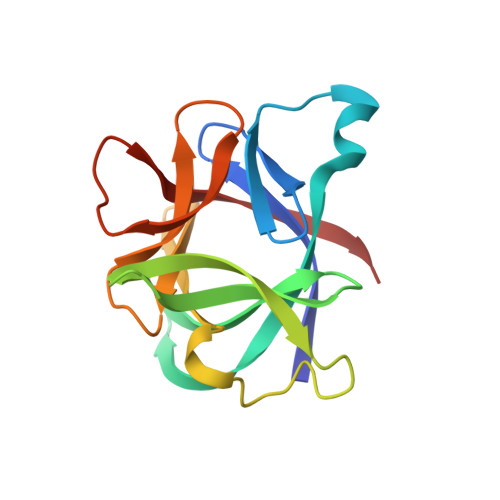
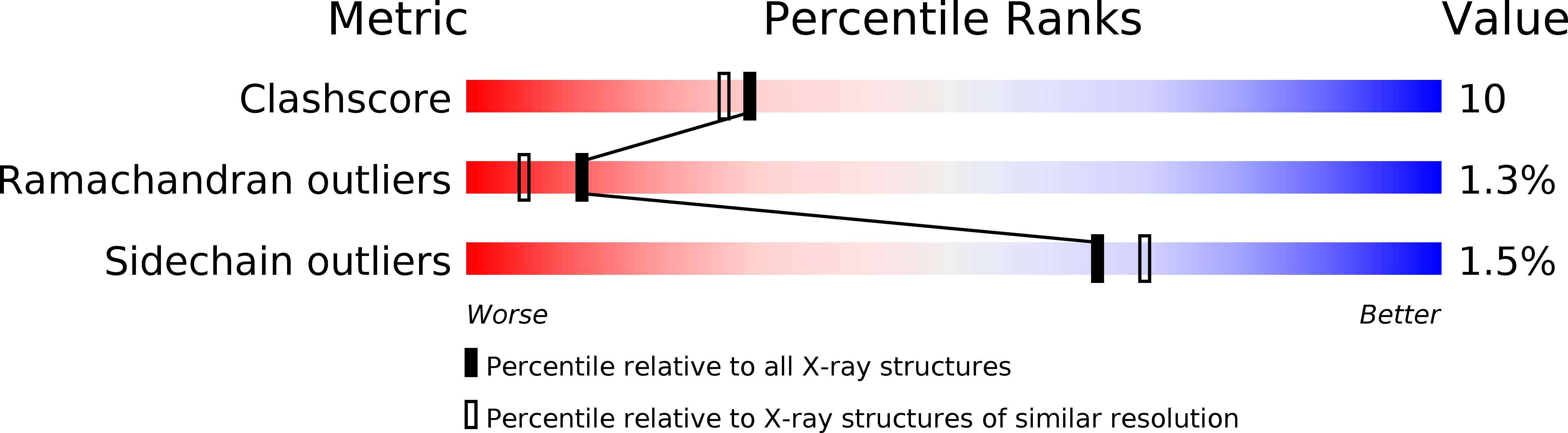Crystal structure of recombinant human interleukin-1 beta at 2.0 A resolution.
Finzel, B.C., Clancy, L.L., Holland, D.R., Muchmore, S.W., Watenpaugh, K.D., Einspahr, H.M.(1989) J Mol Biol 209: 779-791
- PubMed: 2585509
- DOI: https://doi.org/10.1016/0022-2836(89)90606-2
- Primary Citation of Related Structures:
1I1B - PubMed Abstract:
The crystal structure of recombinant human interleukin-1 beta (IL-1 beta) has been determined at 2.0 A resolution and refined to a crystallographic R-factor of 0.19. Three heavy-atom derivatives were identified and used for multiple isomorphous replacement phasing. Interpretation of the resulting electron density map revealed a structure in which there are 12 antiparallel beta-strands and no alpha-helix. The single 153-residue polypeptide chain is folded into a six-stranded beta-barrel similar in architecture to the Kunitz-type trypsin inhibitor found in soybeans. The molecule displays approximate 3-fold symmetry about the axis of the beta-barrel. Each successive pair of component strands of the barrel brackets an extensive sequence outside the barrel that includes an additional pair of beta-strands and a prominent loop. Together, these three external segments conceal much of the perimeter and one end of the barrel, leaving only the end supporting the chain termini fully exposed. The structure can be used to identify portions of the polypeptide chain that are exposed on the surface of the molecule, some of which must be epitopes recognized by interleukin-1 beta receptors.
Organizational Affiliation:
Physical and Analytical Chemistry, Upjohn Company, Kalamazoo, MI 49001.