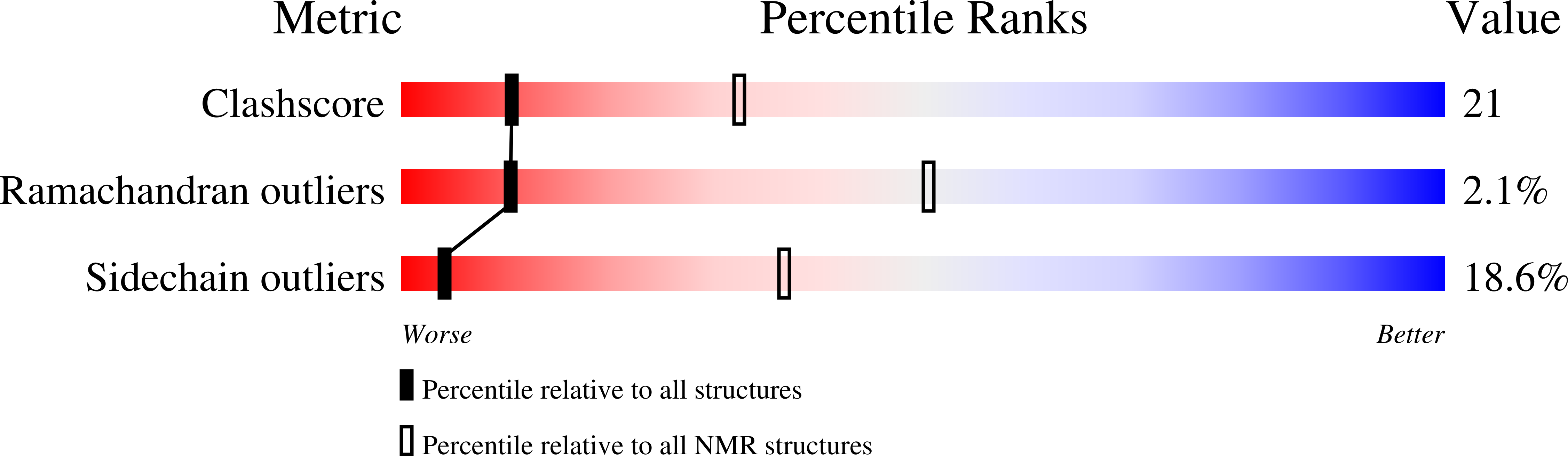The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences.
Freund, C., Dotsch, V., Nishizawa, K., Reinherz, E.L., Wagner, G.(1999) Nat Struct Biol 6: 656-660
- PubMed: 10404223
- DOI: https://doi.org/10.1038/10712
- Primary Citation of Related Structures:
1GYF - PubMed Abstract:
T cell activation through the CD2 cell surface receptor is transmitted by proline-rich sequences within its cytoplasmic tail. A membrane-proximal proline-rich tandem repeat, involved in cytokine production, is recognized by the intracellular CD2 binding protein CD2BP2. We solved the solution structure of the CD2 binding domain of CD2BP2, which we name the glycine-tyrosine-phenylalanine (GYF) domain. The GYF sequence is part of a structurally unique bulge-helix-bulge motif that constitutes the major binding site for the CD2 tail. A hydrophobic surface patch is created by motif residues that are highly conserved among a variety of proteins from diverse eukaryotic species. Thus, the architecture of the GYF domain may be widely used in protein-protein associations.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, Massachusetts 02115, USA. cfreund@wagnerlab.med.harvard.edu