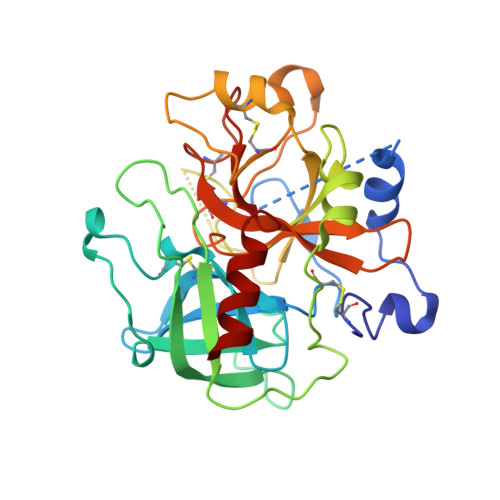
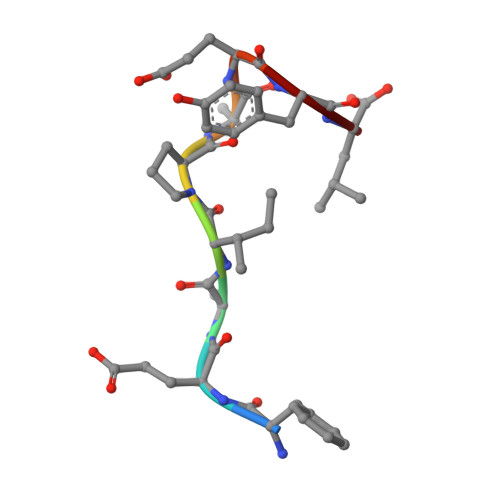
Potent and selective bicyclic lactam inhibitors of thrombin. Part 4: transition state inhibitors.
Bachand, B., Tarazi, M., St-Denis, Y., Edmunds, J.J., Winocour, P.D., Leblond, L., Siddiqui, M.A.(2001) Bioorg Med Chem Lett 11: 287-290
- PubMed: 11212093
- DOI: https://doi.org/10.1016/s0960-894x(00)00636-3
- Primary Citation of Related Structures:
1G37 - PubMed Abstract:
Bicyclic piperazinone based thrombin inhibitors of general structure 2 were prepared and evaluated in vitro and in vivo. These inhibitors, having in common an electrophilic basic trans-cyclohexylamine P1 residue, displayed high thrombin affinity, high selectivity against trypsin and good in vivo efficacy in the rat arterial thrombosis model.
Organizational Affiliation:
BioChem Pharma Inc., Laval, Québec, Canada. bachandb@biochempharma.com