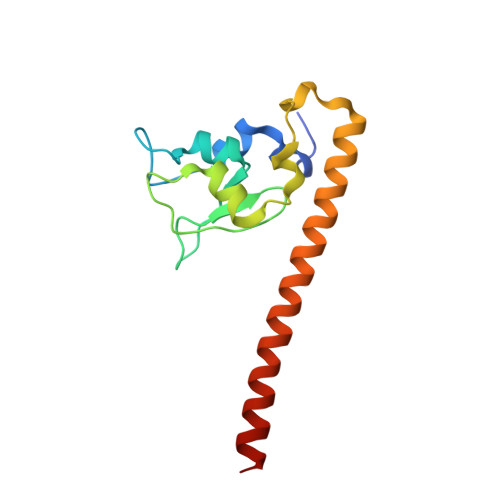
Crystal Structure of Human Survivin Reveals a Bow Tie-Shaped Dimer with Two Unusual Alpha-Helical Extensions
Chantalat, L., Skoufias, D.A., Kleman, J.P., Jung, B., Dideberg, O., Margolis, R.L.(2000) Mol Cell 6: 183
- PubMed: 10949039
- Primary Citation of Related Structures:
1E31 - PubMed Abstract:
Survivin is a mitotic spindle-associated protein involved in linking mitotic spindle function to activation of apoptosis in mammalian cells. The structure of the full-length human survivin has been determined by X-ray crystallography to 2.7 A. Strikingly, the structure forms a very unusual bow tie-shaped dimer. It does not dimerize through a C-terminal coiled-coil, contrary to sequence analysis prediction. The C-terminal helices contain hydrophobic clusters with the potential for protein-protein interactions. The unusual shape and dimensions of survivin suggest it serves an adaptor function through its alpha-helical extensions.
Organizational Affiliation:
Laboratoire de Cristallographie Macromoléculaire, Institut de Biologie Structurale Jean-Pierre Ebel, CEA-CNRS, Grenoble, France.