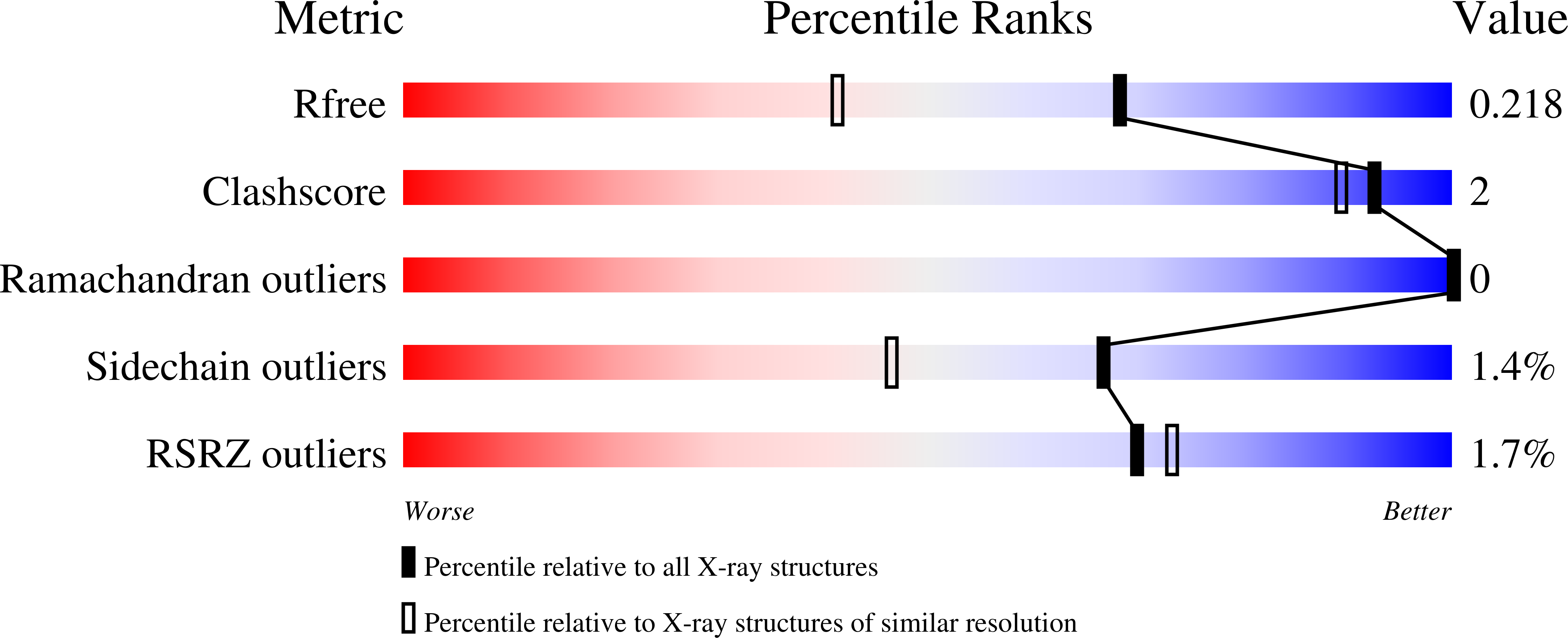
Antibacterial drug leads: DNA and enzyme multitargeting.
Zhu, W., Wang, Y., Li, K., Gao, J., Huang, C.H., Chen, C.C., Ko, T.P., Zhang, Y., Guo, R.T., Oldfield, E.(2015) J Med Chem 58: 1215-1227
- PubMed: 25574764
- DOI: https://doi.org/10.1021/jm501449u
- Primary Citation of Related Structures:
3WYI, 3WYJ, 4U82, 4U8A, 4U8B, 4U8C - PubMed Abstract:
We report the results of an investigation of the activity of a series of amidine and bisamidine compounds against Staphylococcus aureus and Escherichia coli. The most active compounds bound to an AT-rich DNA dodecamer (CGCGAATTCGCG)2 and using DSC were found to increase the melting transition by up to 24 °C. Several compounds also inhibited undecaprenyl diphosphate synthase (UPPS) with IC50 values of 100-500 nM, and we found good correlations (R(2) = 0.89, S. aureus; R(2) = 0.79, E. coli) between experimental and predicted cell growth inhibition by using DNA ΔTm and UPPS IC50 experimental results together with one computed descriptor. We also solved the structures of three bisamidines binding to DNA as well as three UPPS structures. Overall, the results are of general interest in the context of the development of resistance-resistant antibiotics that involve multitargeting.
Organizational Affiliation:
Department of Chemistry, University of Illinois at Urbana-Champaign , 600 South Mathews Avenue, Urbana, Illinois 61801, United States.