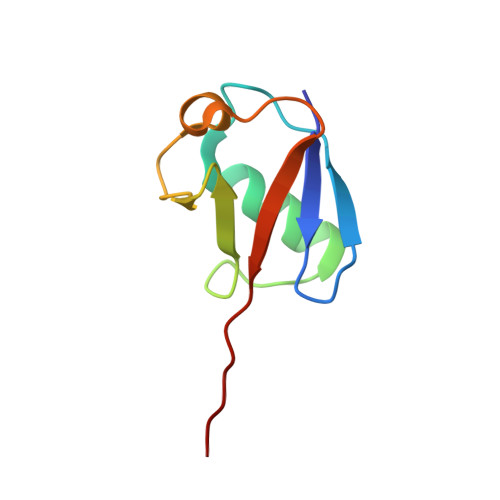
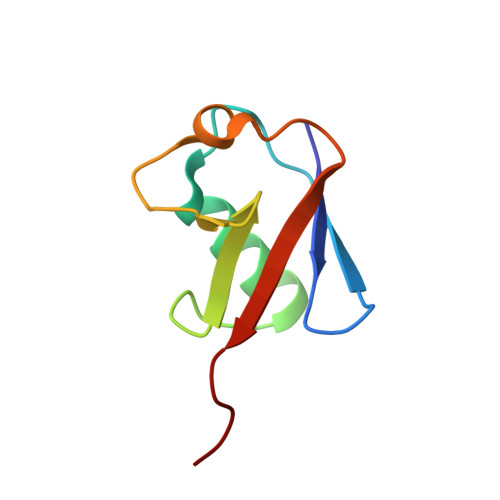
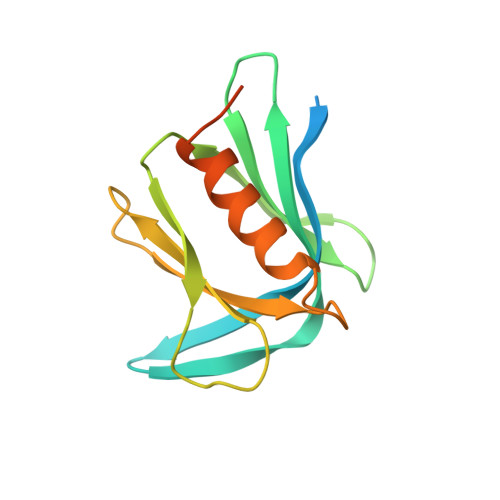
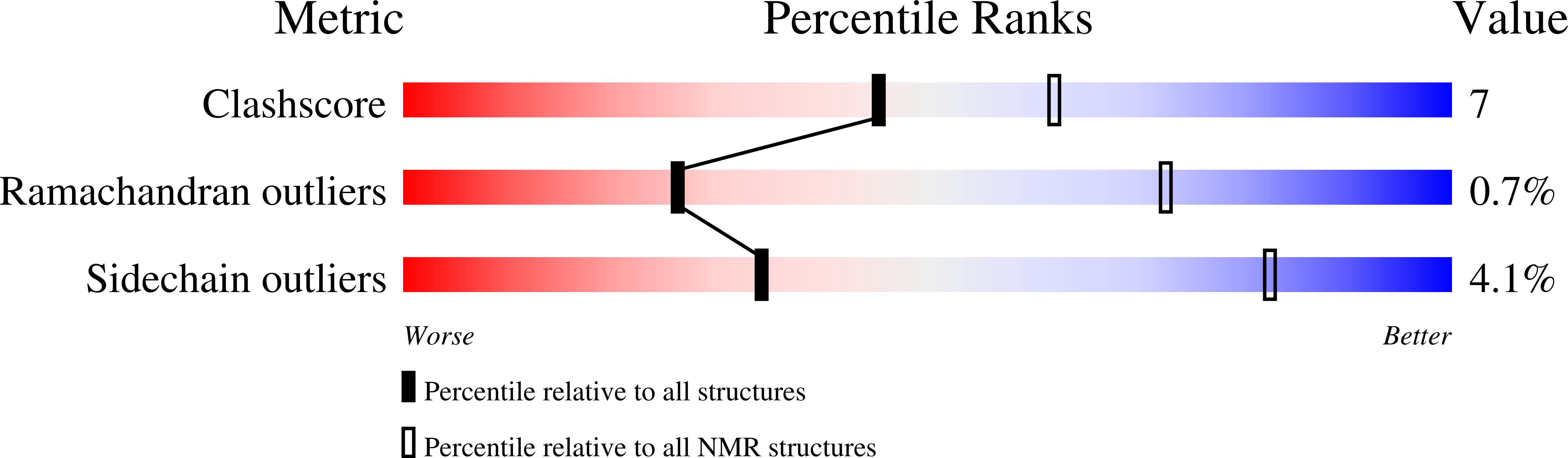An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Lu, X., Ebelle, D.L., Matsuo, H., Walters, K.J.(2020) Structure 28: 495
- PubMed: 32160516
- DOI: https://doi.org/10.1016/j.str.2020.02.007
- Primary Citation of Related Structures:
6UYI, 6UYJ - PubMed Abstract:
Rpn13/Adrm1 is recruited to the proteasome by PSMD1/Rpn2, where it serves as a substrate receptor that binds preferentially to K48-linked ubiquitin chains, an established signal for protein proteolysis. Here, we use NMR to solve the structure of hRpn13 Pru:hRpn2 (940-953):K48-diubiquitin. Surprisingly, hRpn2-bound hRpn13 selects a dynamic, extended conformation of K48-diubiquitin that is unique from previously determined structures. NMR experiments on free K48-diubiquitin demonstrate the presence of the reported "closed" conformation observed by crystallography, but also this more extended state, in which the hRpn13-binding surface is exposed. This extended K48-diubiquitin conformation is defined by interactions between L73 from G76-linked (distal) ubiquitin and a Y59-centered surface of K48-linked (proximal) ubiquitin. Furthermore, hRpn13 exchanges between the two ubiquitins within 100 ms, although prefers the proximal ubiquitin due to interactions with the K48 linker region. Altogether, these data lead to a revised model of how ubiquitinated substrates interact with the proteasome.
Organizational Affiliation:
Protein Processing Section, Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.