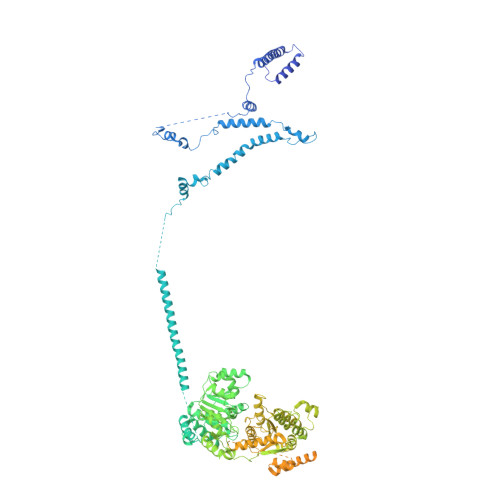
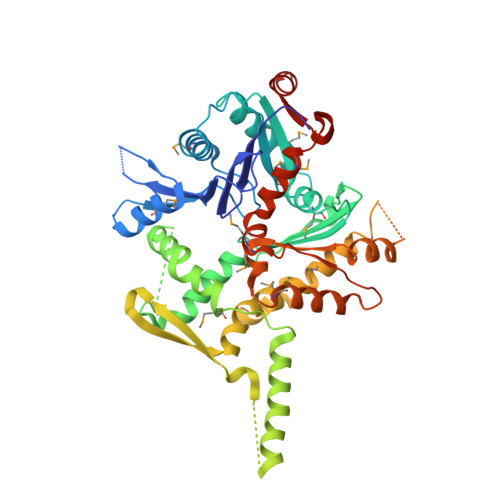
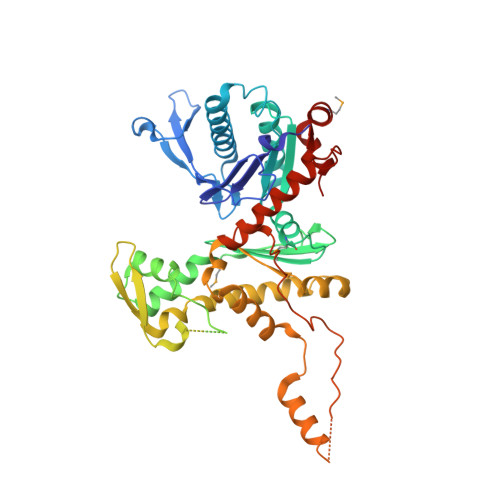
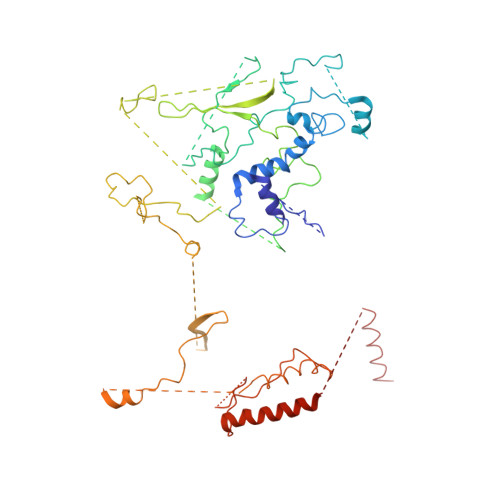
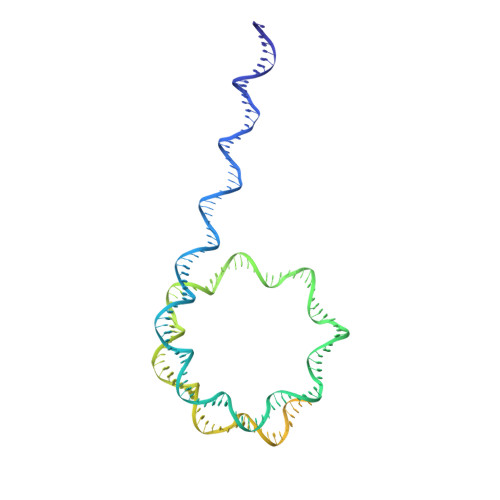
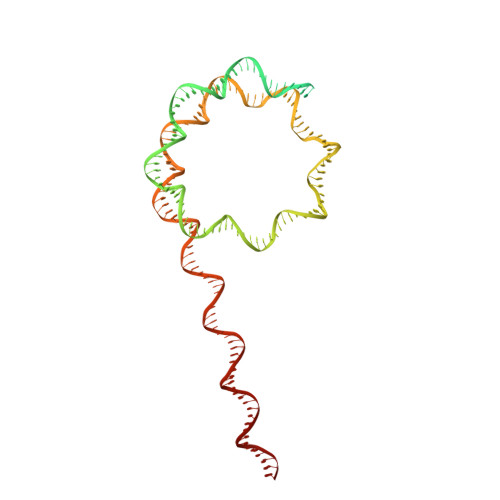
Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Wagner, F.R., Dienemann, C., Wang, H., Stutzer, A., Tegunov, D., Urlaub, H., Cramer, P.(2020) Nature 579: 448-451
- PubMed: 32188943
- DOI: https://doi.org/10.1038/s41586-020-2088-0
- Primary Citation of Related Structures:
6TDA - PubMed Abstract:
Chromatin-remodelling complexes of the SWI/SNF family function in the formation of nucleosome-depleted, transcriptionally active promoter regions (NDRs) 1,2 . In the yeast Saccharomyces cerevisiae, the essential SWI/SNF complex RSC 3 contains 16 subunits, including the ATP-dependent DNA translocase Sth1 4,5 . RSC removes nucleosomes from promoter regions 6,7 and positions the specialized +1 and -1 nucleosomes that flank NDRs 8,9 . Here we present the cryo-electron microscopy structure of RSC in complex with a nucleosome substrate. The structure reveals that RSC forms five protein modules and suggests key features of the remodelling mechanism. The body module serves as a scaffold for the four flexible modules that we call DNA-interacting, ATPase, arm and actin-related protein (ARP) modules. The DNA-interacting module binds extra-nucleosomal DNA and is involved in the recognition of promoter DNA elements 8,10,11 that influence RSC functionality 12 . The ATPase and arm modules sandwich the nucleosome disc with the Snf2 ATP-coupling (SnAC) domain and the finger helix, respectively. The translocase motor of the ATPase module engages with the edge of the nucleosome at superhelical location +2. The mobile ARP module may modulate translocase-nucleosome interactions to regulate RSC activity 5 . The RSC-nucleosome structure provides a basis for understanding NDR formation and the structure and function of human SWI/SNF complexes that are frequently mutated in cancer 13 .
Organizational Affiliation:
Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.