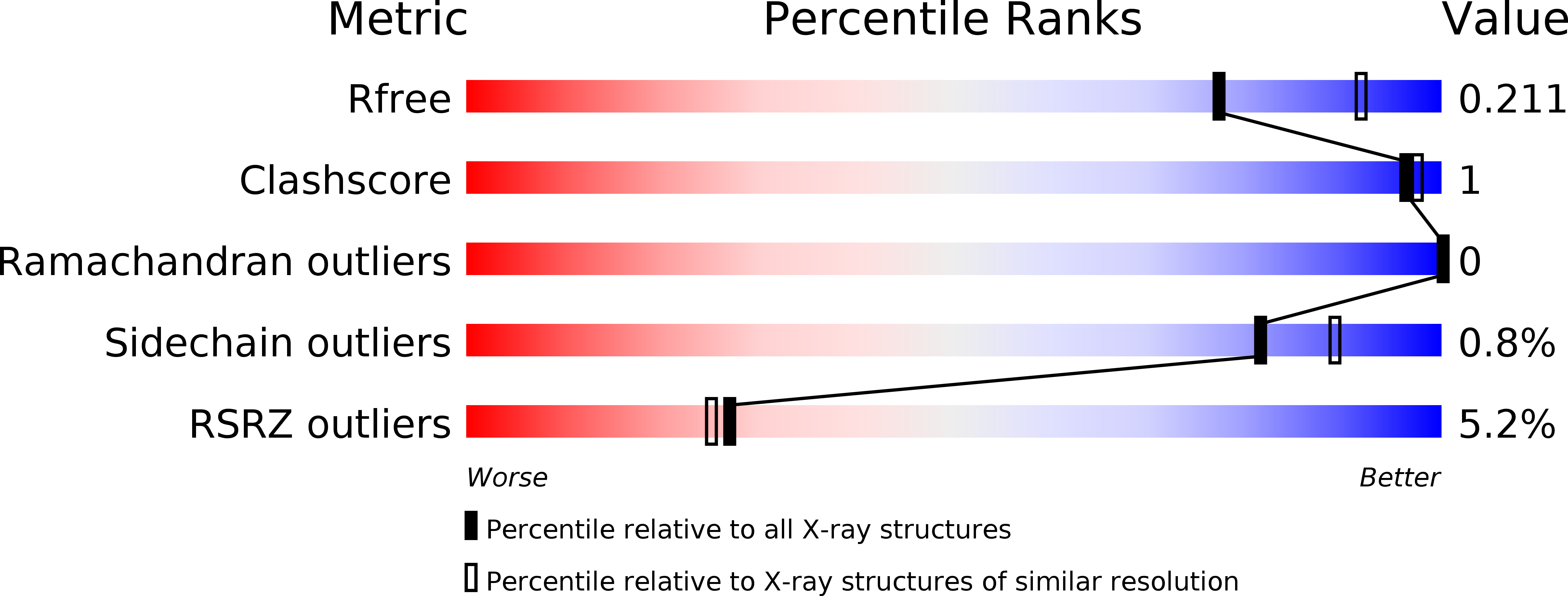
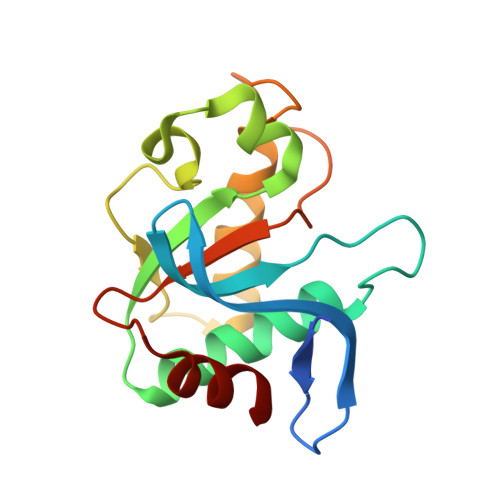
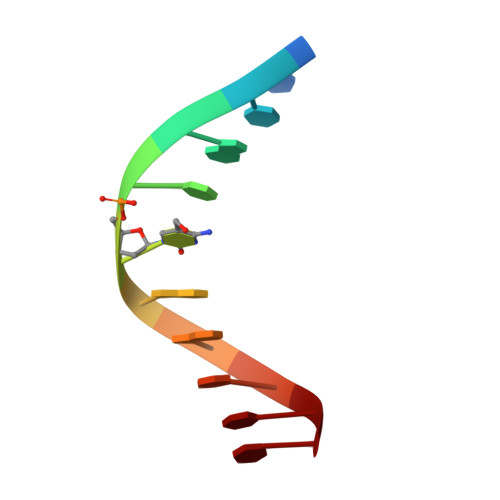
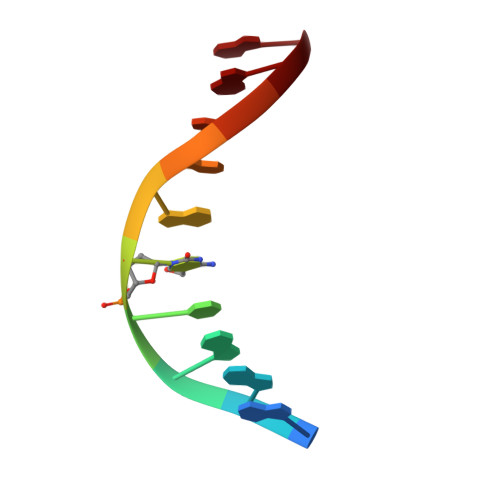
Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Slyvka, A., Zagorskaite, E., Czapinska, H., Sasnauskas, G., Bochtler, M.(2019) Nucleic Acids Res 47: 11943-11955
- PubMed: 31724709
- DOI: https://doi.org/10.1093/nar/gkz1017
- Primary Citation of Related Structures:
6R64, 6T21, 6T22 - PubMed Abstract:
EcoKMcrA from Escherichia coli restricts CpG methylated or hydroxymethylated DNA, and may act as a barrier against host DNA. The enzyme consists of a novel N-terminal specificity domain that we term NEco, and a C-terminal catalytic HNH domain. Here, we report that NEco and full-length EcoKMcrA specificities are consistent. NEco affinity to DNA increases more from hemi- to full-methylation than from non- to hemi-methylation, indicating cooperative binding of the methyl groups. We determined the crystal structures of NEco in complex with fully modified DNA containing three variants of the Y5mCGR EcoKMcrA target sequence: C5mCGG, T5mCGA and T5hmCGA. The structures explain the specificity for the two central base pairs and one of the flanking pairs. As predicted based on earlier biochemical experiments, NEco does not flip any DNA bases. The proximal and distal methyl groups are accommodated in separate pockets. Changes to either pocket reduce DNA binding by NEco and restriction by EcoKMcrA, confirming the relevance of the crystallographically observed binding mode in solution.
Organizational Affiliation:
International Institute of Molecular and Cell Biology, Trojdena 4, 02-109 Warsaw, Poland.