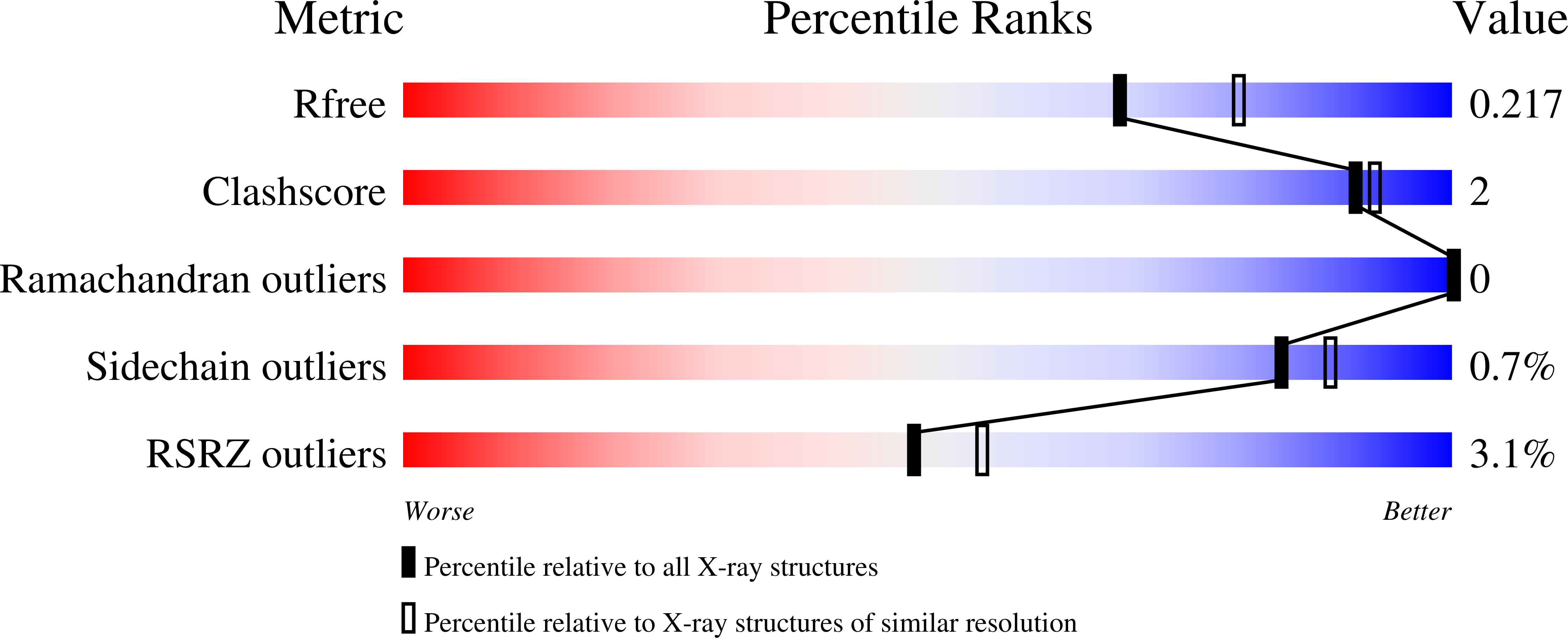
Structural characterization of a short-chain dehydrogenase/reductase from multi-drug resistant Acinetobacter baumannii.
Cross, E.M., Aragao, D., Smith, K.M., Shaw, K.I., Nanson, J.D., Raidal, S.R., Forwood, J.K.(2019) Biochem Biophys Res Commun 518: 465-471
- PubMed: 31443964
- DOI: https://doi.org/10.1016/j.bbrc.2019.08.056
- Primary Citation of Related Structures:
6PZM, 6PZN - PubMed Abstract:
Acinetobacter baumannii (A. baumannii) is a clinically relevant, highly drug-resistant pathogen of global concern. An attractive approach to drug design is to specifically target the type II fatty acid synthesis (FASII) pathway which is critical in Gram negative bacteria and is significantly different to the type I fatty acid synthesis (FASI) pathway found in mammals. Enzymes involved in FASII include members of the short-chain dehydrogenase/reductase (SDR) superfamily. SDRs are capable of performing a diverse range of biochemical reactions against a broad spectrum of substrates whilst maintaining conserved structural features and sequence motifs. Here, we use X-ray crystallography to describe the structure of an SDR from the multi-drug resistant bacteria A. baumannii, previously annotated as a putative FASII FabG enzyme. The protein was recombinantly expressed, purified, and crystallized. The protein crystals diffracted to 2.0 Å and the structure revealed a FabG-like fold. Functional assays revealed, however, that the protein was not active against the FabG substrate, acetoacetyl-CoA. This study highlights that database annotations may show the necessary structural hallmarks of such proteins, however, they may not be able to cleave substrates that are typical of FabG enzymes. These results are important for the selection of target enzymes in future drug development.
Organizational Affiliation:
School of Biomedical Sciences, Charles Sturt University, Wagga Wagga, NSW, 2678, Australia.