Inhibition of Marburg Virus RNA Synthesis by a Synthetic Anti-VP35 Antibody.
Amatya, P., Wagner, N., Chen, G., Luthra, P., Shi, L., Borek, D., Pavlenco, A., Rohrs, H., Basler, C.F., Sidhu, S.S., Gross, M.L., Leung, D.W.(2019) ACS Infect Dis 5: 1385-1396
- PubMed: 31120240
- DOI: https://doi.org/10.1021/acsinfecdis.9b00091
- Primary Citation of Related Structures:
6OTC - PubMed Abstract:
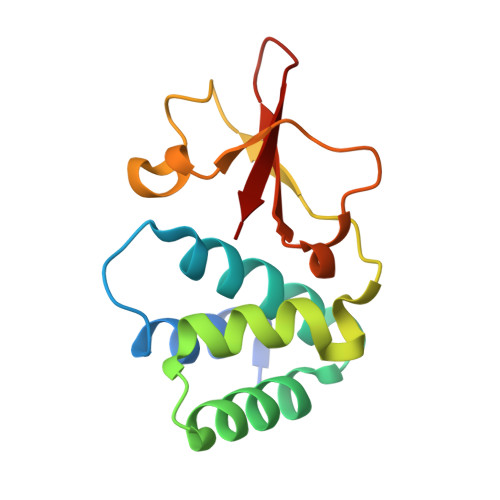
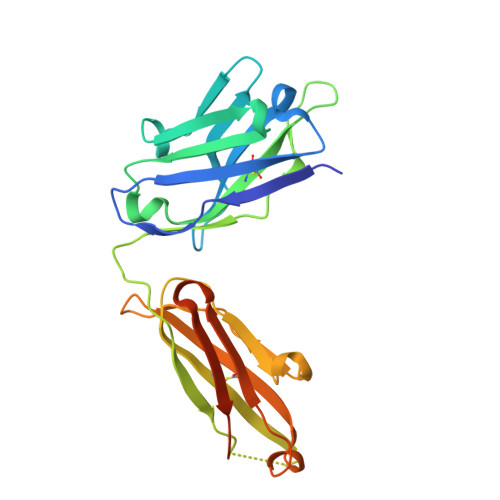
Marburg virus causes sporadic outbreaks of severe hemorrhagic fever with high case fatality rates. Approved, effective, and safe therapeutic or prophylactic countermeasures are lacking. To address this, we used phage display to engineer a synthetic antibody, sFab H3, which binds the Marburg virus VP35 protein (mVP35). mVP35 is a critical cofactor of the viral replication complex and a viral immune antagonist. sFab H3 displayed high specificity for mVP35 and not for the closely related Ebola virus VP35. sFab H3 inhibited viral-RNA synthesis in a minigenome assay, suggesting its potential use as an antiviral. We characterized sFab H3 by a combination of biophysical and biochemical methods, and a crystal structure of the complex solved to 1.7 Å resolution defined the molecular interface between the sFab H3 and mVP35 interferon inhibitory domain. Our study identifies mVP35 as a therapeutic target using an approach that provides a framework for generating engineered Fabs targeting other viral proteins.
Organizational Affiliation:
Department of Medicine , Washington University School of Medicine , 660 South Euclid Avenue , St. Louis , Missouri 63110 , United States.