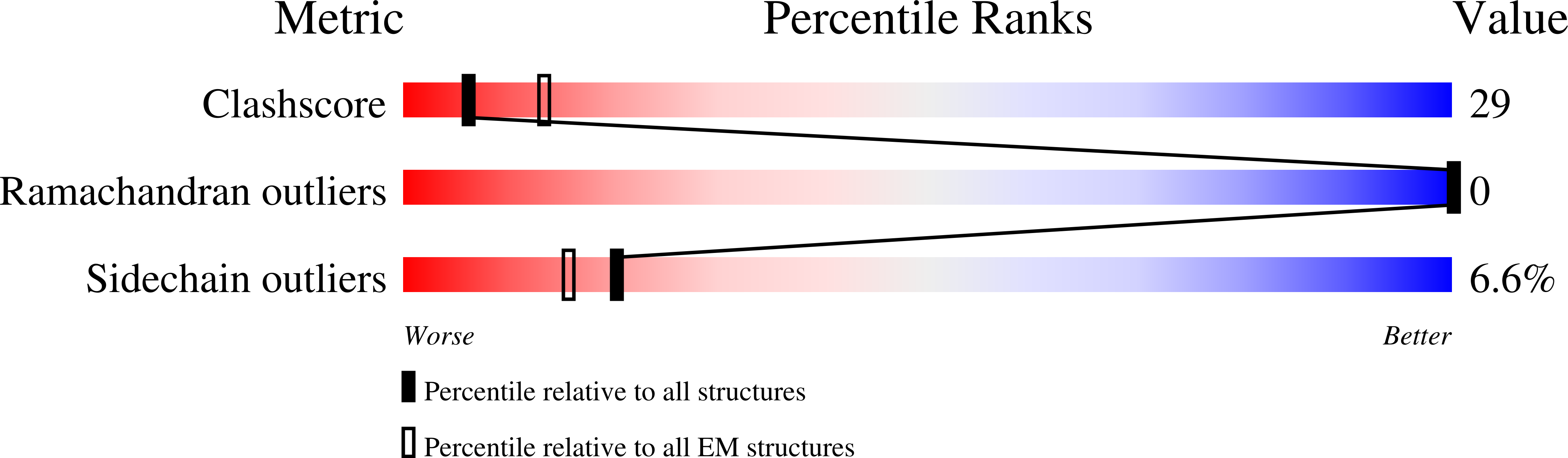Structural Insights into alpha-Synuclein Fibril Polymorphism: Effects of Parkinson's Disease-Related C-Terminal Truncations.
Ni, X., McGlinchey, R.P., Jiang, J., Lee, J.C.(2019) J Mol Biol 431: 3913-3919
- PubMed: 31295458
- DOI: https://doi.org/10.1016/j.jmb.2019.07.001
- Primary Citation of Related Structures:
6OSJ, 6OSL, 6OSM - PubMed Abstract:
Lewy bodies, hallmarks of Parkinson's disease, contain C-terminally truncated (ΔC) α-synuclein (α-syn). Here, we report fibril structures of three N-terminally acetylated (Ac) α-syn constructs, Ac1-140, Ac1-122, and Ac1-103, solved by cryoelectron microscopy. Both ΔC-α-syn variants exhibited faster aggregation kinetics, and Ac1-103 fibrils efficiently seeded the full-length protein, highlighting their importance in pathogenesis. Interestingly, fibril helical twists increased upon the removal of C-terminal residues and can be propagated through cross-seeding. Compared to that of Ac1-140, increased electron densities were seen in the N-terminus of Ac1-103, whereas the C-terminus of Ac1-122 appeared more structured. In accord, the respective termini of ΔC-α-syn exhibited increased protease resistance. Despite similar amyloid core residues, distinctive features were seen for both Ac1-122 and Ac1-103. Particularly, Ac1-103 has the tightest packed core with an additional turn, likely attributable to conformational changes in the N-terminal region. These molecular differences offer insights into the effect of C-terminal truncations on α-syn fibril polymorphism.
Organizational Affiliation:
Laboratory of Membrane Proteins and Structural Biology, Biochemistry and Biophysics Center, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892, USA.