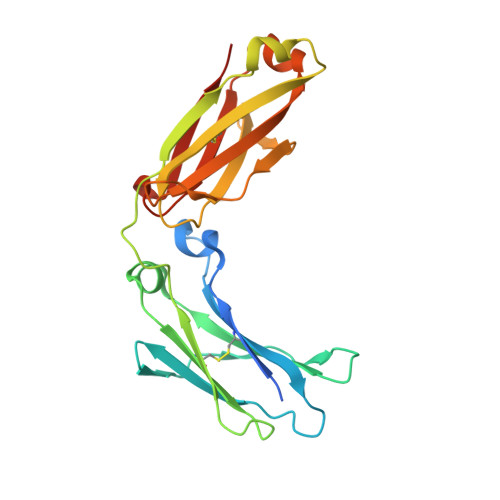
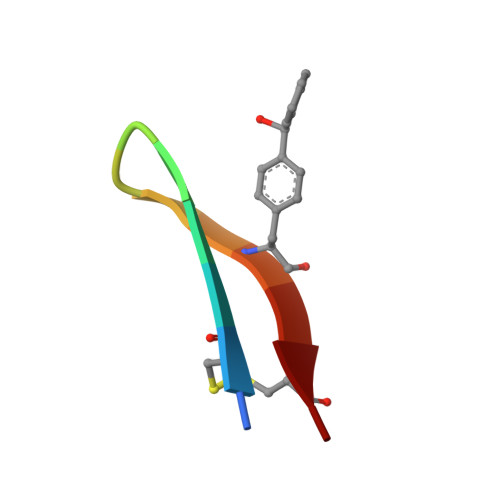
Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Vance, N., Zacharias, N., Ultsch, M., Li, G., Fourie, A., Liu, P., LaFrance-Vanasse, J., Ernst, J.A., Sandoval, W., Kozak, K.R., Phillips, G., Wang, W., Sadowsky, J.(2019) Bioconjug Chem 30: 148-160
- PubMed: 30566343
- DOI: https://doi.org/10.1021/acs.bioconjchem.8b00809
- Primary Citation of Related Structures:
6N9T - PubMed Abstract:
Site-specific conjugation of small molecules to antibodies represents an attractive goal for the development of more homogeneous targeted therapies and diagnostics. Most site-specific conjugation strategies require modification or removal of antibody glycans or interchain disulfide bonds or engineering of an antibody mutant that bears a reactive handle. While such methods are effective, they complicate the process of preparing antibody conjugates and can negatively impact biological activity. Herein we report the development and detailed characterization of a robust photoaffinity cross-linking method for site-specific conjugation to fully glycosylated wild-type antibodies. The method employs a benzoylphenylalanine (Bpa) mutant of a previously described 13-residue peptide derived from phage display to bind tightly to the Fc domain; upon UV irradiation, the Bpa residue forms a diradical that reacts with the bound antibody. After the initial discovery of an effective Bpa mutant peptide and optimization of the reaction conditions to enable efficient conjugation without concomitant UV-induced photodamage of the antibody, we assessed the scope of the photoconjugation reaction across different human and nonhuman antibodies and antibody mutants. Next, the specific site of conjugation on a human antibody was characterized in detail by mass spectrometry experiments and at atomic resolution by X-ray crystallography. Finally, we adapted the photoconjugation method to attach a cytotoxic payload site-specifically to a wild-type antibody and showed that the resulting conjugate is both stable in plasma and as potent as a conventional antibody-drug conjugate in cells, portending well for future biological applications.
Organizational Affiliation:
Research & Early Development , Genentech, Inc. , 1 DNA Way , South San Francisco , California 94080 , United States.