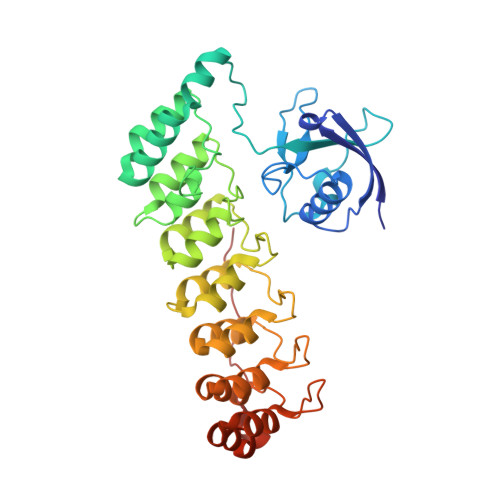
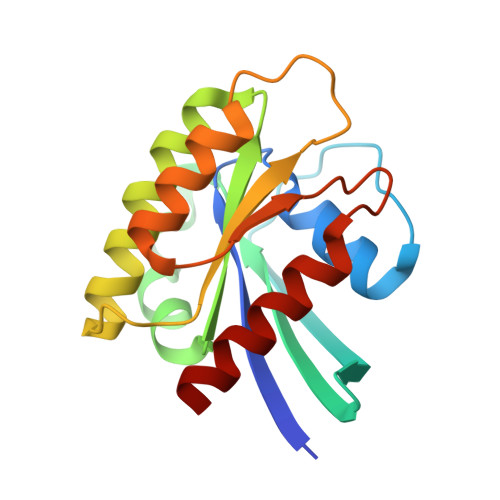
Shank3 Binds to and Stabilizes the Active Form of Rap1 and HRas GTPases via Its NTD-ANK Tandem with Distinct Mechanisms.
Cai, Q., Hosokawa, T., Zeng, M., Hayashi, Y., Zhang, M.(2020) Structure 28: 290
- PubMed: 31879129
- DOI: https://doi.org/10.1016/j.str.2019.11.018
- Primary Citation of Related Structures:
6KYH, 6KYK - PubMed Abstract:
Shank1/2/3, major scaffold proteins in excitatory synapses, are frequently mutated in patients with psychiatric disorders. Although the Shank N-terminal domain and ankyrin repeats domain tandem (NTD-ANK) is known to bind to Ras and Rap1, the molecular mechanism underlying and functional significance of the bindings in synapses are unknown. Here, we demonstrate that Shank3 NTD-ANK specifically binds to the guanosine triphosphate (GTP)-bound form of HRas and Rap1. In addition to the canonical site mediated by the Ras-association domain and common to both GTPases, Shank3 contains an unconventional Rap1 binding site formed by NTD and ANK together. Binding of Shank3 to the GTP-loaded Rap1 slows down its GTP hydrolysis by SynGAP. We further show that the interactions between Shank3 and HRas/Rap1 at excitatory synapses are promoted by synaptic activation. Thus, Shank3 may be able to modulate signaling of the Ras family proteins via directly binding to and stabilizing the GTP-bound form of the enzymes.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong, China.