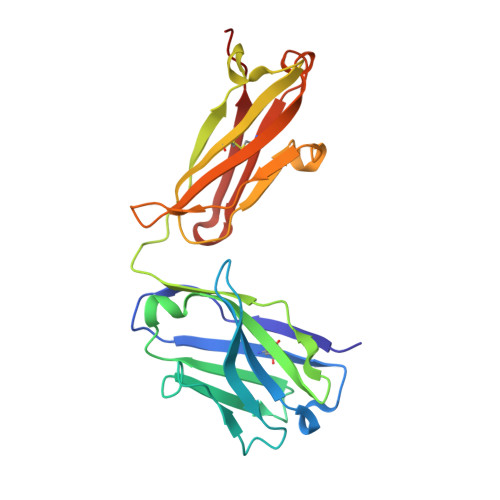
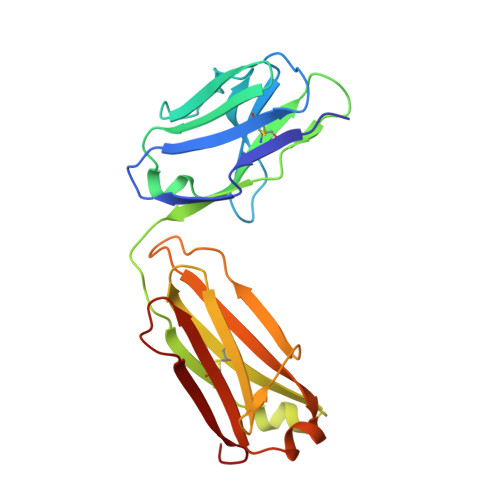
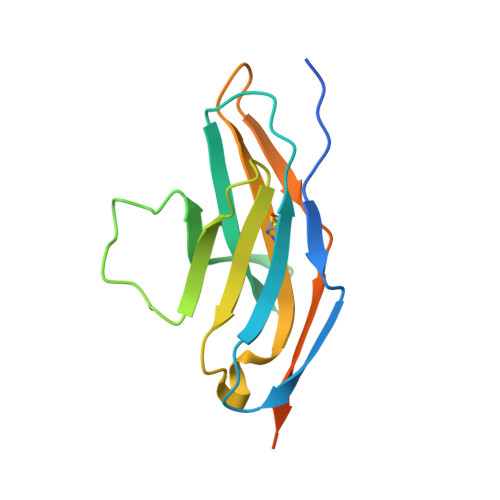
Identification of a monoclonal antibody that targets PD-1 in a manner requiring PD-1 Asn58 glycosylation.
Wang, M., Wang, J., Wang, R., Jiao, S., Wang, S., Zhang, J., Zhang, M.(2019) Commun Biol 2: 392-392
- PubMed: 31667366
- DOI: https://doi.org/10.1038/s42003-019-0642-9
- Primary Citation of Related Structures:
6JJP - PubMed Abstract:
Programmed cell death 1 (PD-1) is inhibitory receptor and immune checkpoint protein. Blocking the interaction of PD-1 and its ligands PD-L1/ L2 is able to active T-cell-mediated antitumor response. Monoclonal antibody-based drugs targeting PD-1 pathway have exhibited great promise in cancer therapy. Here we show that MW11-h317, an anti-PD-1 monoclonal antibody, displays high affinity for PD-1 and blocks PD-1 interactions with PD-L1/L2. MW11-h317 can effectively induce T-cell-mediated immune response and inhibit tumor growth in mouse model. Crystal structure of PD-1/MW11-h317 Fab complex reveals that both the loops and glycosylation of PD-1 are involved in recognition and binding, in which Asn58 glycosylation plays a critical role. The unique glycan epitope in PD-1 to MW11-h317 is different from the first two approved clinical PD-1 antibodies, nivolumab and pembrolizumab. These results suggest MW11-h317 as a therapeutic monoclonal antibody of PD-1 glycosylation-targeting which may become efficient alternative for cancer therapy.
Organizational Affiliation:
1School of Life Sciences, Anhui University, 230601 Hefei, Anhui China.