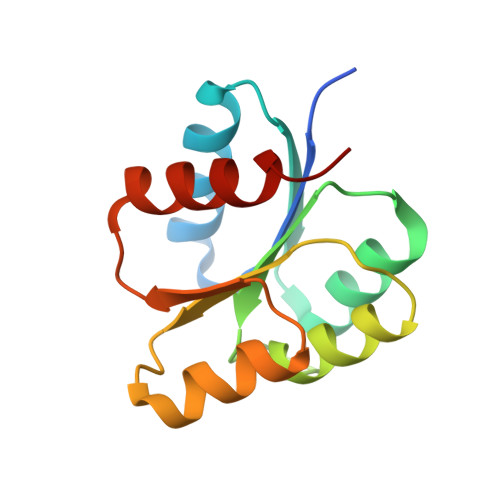
Deciphering the activation and recognition mechanisms of Staphylococcus aureus response regulator ArlR.
Ouyang, Z., Zheng, F., Chew, J.Y., Pei, Y., Zhou, J., Wen, K., Han, M., Lemieux, M.J., Hwang, P.M., Wen, Y.(2019) Nucleic Acids Res 47: 11418-11429
- PubMed: 31598698
- DOI: https://doi.org/10.1093/nar/gkz891
- Primary Citation of Related Structures:
6IS1, 6IS2, 6IS3, 6IS4 - PubMed Abstract:
Staphylococcus aureus ArlRS is a key two-component regulatory system necessary for adhesion, biofilm formation, and virulence. The response regulator ArlR consists of a C-terminal DNA-binding effector domain and an N-terminal receiver domain that is phosphorylated by ArlS, the cognate transmembrane sensor histidine kinase. We demonstrate that the receiver domain of ArlR adopts the canonical α5β5 response regulator assembly, which dimerizes upon activation, using beryllium trifluoride as an aspartate phosphorylation mimic. Activated ArlR recognizes a 20-bp imperfect inverted repeat sequence in the ica operon, which is involved in intercellular adhesion polysaccharide production. Crystal structures of the inactive and activated forms reveal that activation induces a significant conformational change in the β4-α4 and β5-α5-connecting loops, in which the α4 and α5 helices constitute the homodimerization interface. Crystal structures of the DNA-binding ArlR effector domain indicate that it is able to dimerize via a non-canonical β1-β2 hairpin domain swapping, raising the possibility of a new mechanism for signal transduction from the receiver domain to effector domain. Taken together, the current study provides structural insights into the activation of ArlR and its recognition, adding to the diversity of response regulation mechanisms that may inspire novel antimicrobial strategies specifically targeting Staphylococcus.
Organizational Affiliation:
The Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Department of Biochemistry and Molecular Biology, The Key Laboratory of Environment and Genes Related to Disease of Ministry of Education, Health Science Center, Xi'an Jiaotong University, Xi'an, China.