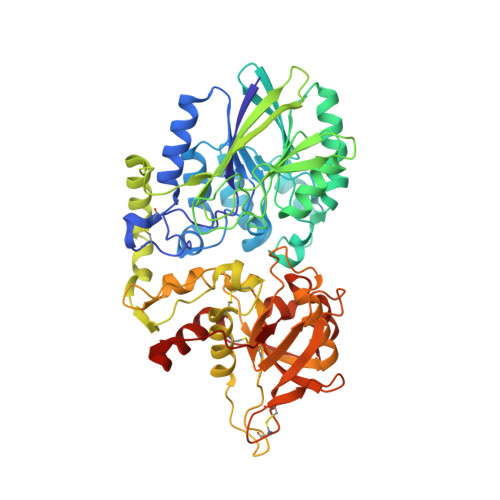
Blocking Antibodies Targeting the CD39/CD73 Immunosuppressive Pathway Unleash Immune Responses in Combination Cancer Therapies.
Perrot, I., Michaud, H.A., Giraudon-Paoli, M., Augier, S., Docquier, A., Gros, L., Courtois, R., Dejou, C., Jecko, D., Becquart, O., Rispaud-Blanc, H., Gauthier, L., Rossi, B., Chanteux, S., Gourdin, N., Amigues, B., Roussel, A., Bensussan, A., Eliaou, J.F., Bastid, J., Romagne, F., Morel, Y., Narni-Mancinelli, E., Vivier, E., Paturel, C., Bonnefoy, N.(2019) Cell Rep 27: 2411-2425.e9
- PubMed: 31116985
- DOI: https://doi.org/10.1016/j.celrep.2019.04.091
- Primary Citation of Related Structures:
6HXW - PubMed Abstract:
Immune checkpoint inhibitors have revolutionized cancer treatment. However, many cancers are resistant to ICIs, and the targeting of additional inhibitory signals is crucial for limiting tumor evasion. The production of adenosine via the sequential activity of CD39 and CD73 ectoenzymes participates to the generation of an immunosuppressive tumor microenvironment. In order to disrupt the adenosine pathway, we generated two antibodies, IPH5201 and IPH5301, targeting human membrane-associated and soluble forms of CD39 and CD73, respectively, and efficiently blocking the hydrolysis of immunogenic ATP into immunosuppressive adenosine. These antibodies promoted antitumor immunity by stimulating dendritic cells and macrophages and by restoring the activation of T cells isolated from cancer patients. In a human CD39 knockin mouse preclinical model, IPH5201 increased the anti-tumor activity of the ATP-inducing chemotherapeutic drug oxaliplatin. These results support the use of anti-CD39 and anti-CD73 monoclonal antibodies and their combination with immune checkpoint inhibitors and chemotherapies in cancer.
Organizational Affiliation:
Innate Pharma, 117 Avenue de Luminy, 13009 Marseille, France.