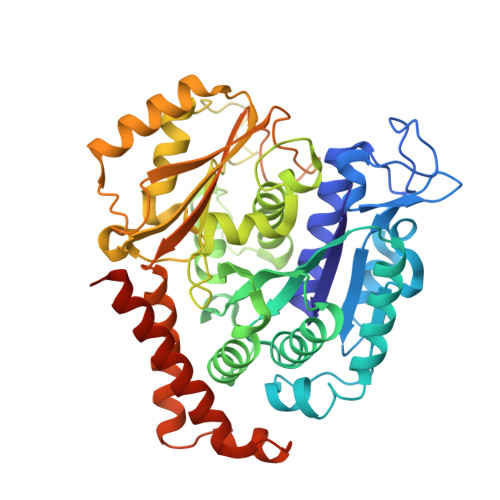
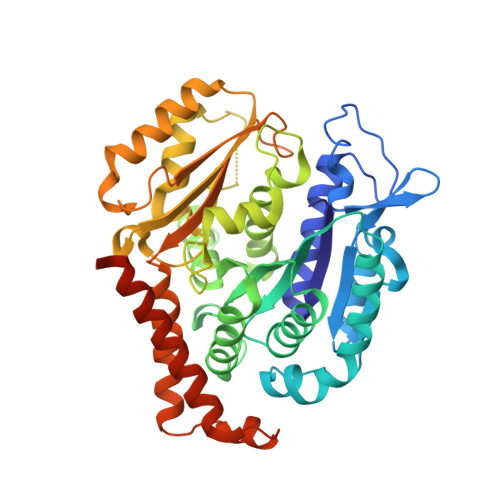
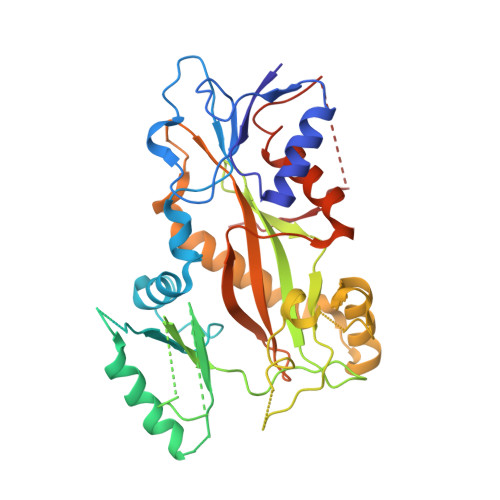
Tetrahydroisoquinoline Sulfamates as Potent Microtubule Disruptors: Synthesis, Antiproliferative and Antitubulin Activity of Dichlorobenzyl-Based Derivatives, and a Tubulin Cocrystal Structure.
Dohle, W., Prota, A.E., Menchon, G., Hamel, E., Steinmetz, M.O., Potter, B.V.L.(2019) ACS Omega 4: 755-764
- PubMed: 30775645
- DOI: https://doi.org/10.1021/acsomega.8b02879
- Primary Citation of Related Structures:
6HX8 - PubMed Abstract:
Tetrahydroisoquinoline (THIQ) 6- O -sulfamate-based anticancer agents, inspired by the endogenous steroid 2-methoxyestradiol and its sulfamate derivatives, are further explored for antiproliferative and microtubule disruptor activity. Based on recently designed C3-methyl C7-methoxy-substituted THIQ derivatives, compounds with mono- and dichloro-substitutions on the pendant N -benzyl ring were synthesized and evaluated. Although improved antiproliferative activity was observed, for example, 4a versus 4b and 4b versus 8c , it was relatively modest. Compound 8c , a 2',5'-dichlorobenzyl derivative was, however, identified as a promising antiproliferative agent with in vitro activities exceeding that of the parent steroid (e.g., GI 50 90 nM in DU-145 cells) and was highly potent against a range of tumor cell lines (e.g., GI 50 26 nM for OVCAR-3). 8c inhibited the polymerization of tubulin in vitro with an IC 50 only twofold less potent than combretastatin A-4 and inhibited colchicine binding to tubulin. Tubulin polymerization assays showed the parent THIQ 4a to be only a very weak inhibitor, but a striking potency difference was seen between compounds with C2' methoxy and chloro substituents, whereas this was much smaller when these substituents were positioned at C5'. To confirm the target in atomic detail and because 8c is a racemic mixture, an achiral parent THIQ 6- O -sulfamate derivative 10 was successfully cocrystallized with the αβ-tubulin heterodimer. The derivative 10 binds at the colchicine site on tubulin, the first example of this compound class investigated in such detail, with its sulfamate group interacting with residues beyond the reach of colchicine itself, similar to a recently reported quinazolinone sulfamate derivative, 6a . The structure also suggests that for racemic C3-methyl-substituted THIQ derivatives, such as 8c , the (S)-enantiomer is likely to be preferentially accommodated within the colchicine site for steric reasons. The results further confirm the potential of nonsteroidal THIQ sulfamate derivatives for oncology and suggest that the mechanism of microtubule destabilization for the THIQ compound class is to prevent the curved-to-straight conformational transition of tubulin required for polymerization.
Organizational Affiliation:
Medicinal Chemistry & Drug Discovery, Department of Pharmacology, University of Oxford, Mansfield Road, Oxford OX1 3QT, U.K.