Discovery of Potent, Selective, and Peripherally Restricted Pan-Trk Kinase Inhibitors for the Treatment of Pain.
Bagal, S.K., Andrews, M., Bechle, B.M., Bian, J., Bilsland, J., Blakemore, D.C., Braganza, J.F., Bungay, P.J., Corbett, M.S., Cronin, C.N., Cui, J.J., Dias, R., Flanagan, N.J., Greasley, S.E., Grimley, R., James, K., Johnson, E., Kitching, L., Kraus, M.L., McAlpine, I., Nagata, A., Ninkovic, S., Omoto, K., Scales, S., Skerratt, S.E., Sun, J., Tran-Dube, M., Waldron, G.J., Wang, F., Warmus, J.S.(2018) J Med Chem 61: 6779-6800
- PubMed: 29944371
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00633
- Primary Citation of Related Structures:
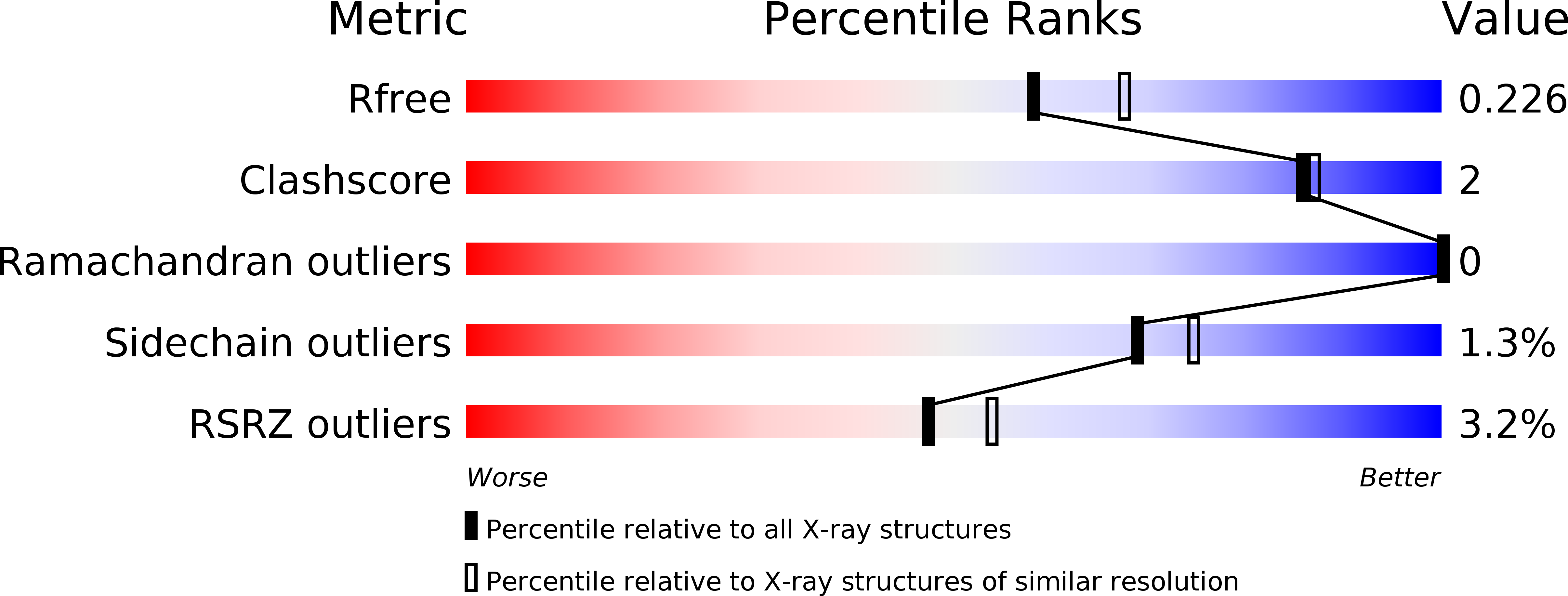
6DKB, 6DKG, 6DKI, 6DKW - PubMed Abstract:
Hormones of the neurotrophin family, nerve growth factor (NGF), brain derived neurotrophic factor (BDNF), neurotrophin 3 (NT3), and neurotrophin 4 (NT4), are known to activate the family of Tropomyosin receptor kinases (TrkA, TrkB, and TrkC). Moreover, inhibition of the TrkA kinase pathway in pain has been clinically validated by the NGF antibody tanezumab, leading to significant interest in the development of small molecule inhibitors of TrkA. Furthermore, Trk inhibitors having an acceptable safety profile will require minimal brain availability. Herein, we discuss the discovery of two potent, selective, peripherally restricted, efficacious, and well-tolerated series of pan-Trk inhibitors which successfully delivered three candidate quality compounds 10b, 13b, and 19. All three compounds are predicted to possess low metabolic clearance in human that does not proceed via aldehyde oxidase-catalyzed reactions, thus addressing the potential clearance prediction liability associated with our current pan-Trk development candidate PF-06273340.
Organizational Affiliation:
Worldwide Medicinal Chemistry , Pfizer Worldwide R&D U.K. , The Portway Building, Granta Park , Cambridge CB21 6GS , U.K.