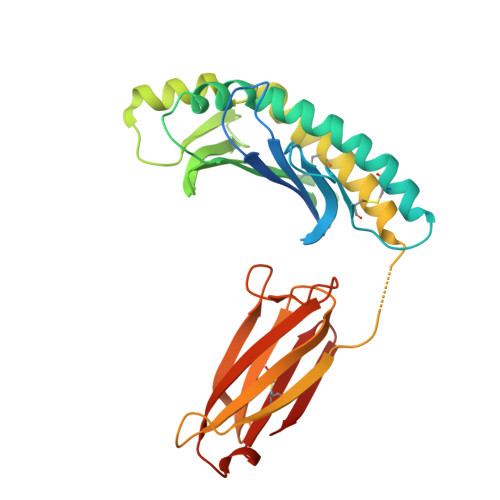
Structure of MHC class I-like MILL2 reveals heparan-sulfate binding and interdomain flexibility.
Kajikawa, M., Ose, T., Fukunaga, Y., Okabe, Y., Matsumoto, N., Yonezawa, K., Shimizu, N., Kollnberger, S., Kasahara, M., Maenaka, K.(2018) Nat Commun 9: 4330-4330
- PubMed: 30337538
- DOI: https://doi.org/10.1038/s41467-018-06797-8
- Primary Citation of Related Structures:
6A97 - PubMed Abstract:
The MILL family, composed of MILL1 and MILL2, is a group of nonclassical MHC class I molecules that occur in some orders of mammals. It has been reported that mouse MILL2 is involved in wound healing; however, the molecular mechanisms remain unknown. Here, we determine the crystal structure of MILL2 at 2.15 Å resolution, revealing an organization similar to classical MHC class I. However, the α1-α2 domains are not tightly fixed on the α3-β 2 m domains, indicating unusual interdomain flexibility. The groove between the two helices in the α1-α2 domains is too narrow to permit ligand binding. Notably, an unusual basic patch on the α3 domain is involved in the binding to heparan sulfate which is essential for MILL2 interactions with fibroblasts. These findings suggest that MILL2 has a unique structural architecture and physiological role, with binding to heparan sulfate proteoglycans on fibroblasts possibly regulating cellular recruitment in biological events.
Organizational Affiliation:
Laboratory of Microbiology, Showa Pharmaceutical University, Machida, Tokyo, 190-8543, Japan.