Engineered promoter selectivity of an ECF sigma factor
Campagne, S., Vorholt, J.A., Allain, F.H.Not Published
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Not Published
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| ECF RNA polymerase sigma-E factor,ECF RNA polymerase sigma factor SigW,ECF RNA polymerase sigma-E factor | 95 | Escherichia coli K-12, Bacillus subtilis subsp. subtilis str. 168 | Mutation(s): 0 Gene Names: rpoE, sigE, b2573, JW2557, sigW, ybbL, BSU01730 | 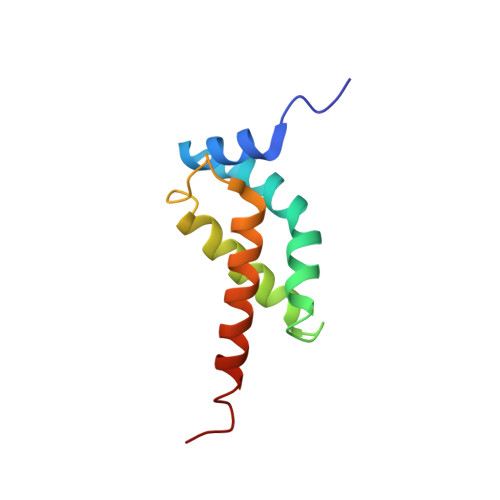 | |
UniProt | |||||
Find proteins for P0AGB6 (Escherichia coli (strain K12)) Explore P0AGB6 Go to UniProtKB: P0AGB6 | |||||
Find proteins for Q45585 (Bacillus subtilis (strain 168)) Explore Q45585 Go to UniProtKB: Q45585 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q45585P0AGB6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*GP*TP*AP*AP*AP*A)-3') | 6 | synthetic construct | 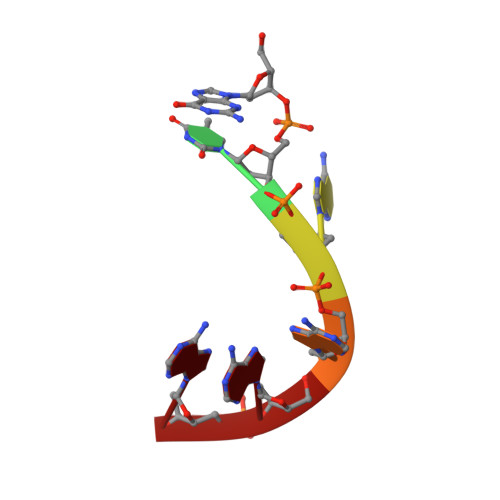 | ||
Sequence AnnotationsExpand | |||||
| |||||