Structure and mechanism of a group-I cobalt energy coupling factor transporter
Bao, Z., Qi, X., Hong, S., Xu, K., He, F., Zhang, M., Chen, J., Chao, D., Zhao, W., Li, D., Wang, J., Zhang, P.(2017) Cell Res 27: 675-687
- PubMed: 28322252
- DOI: https://doi.org/10.1038/cr.2017.38
- Primary Citation of Related Structures:
5X3X, 5X40, 5X41 - PubMed Abstract:
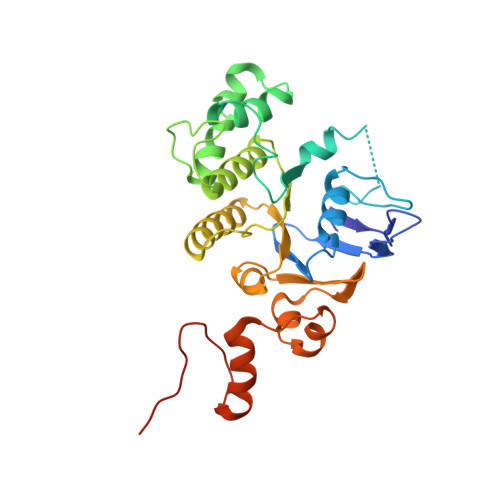
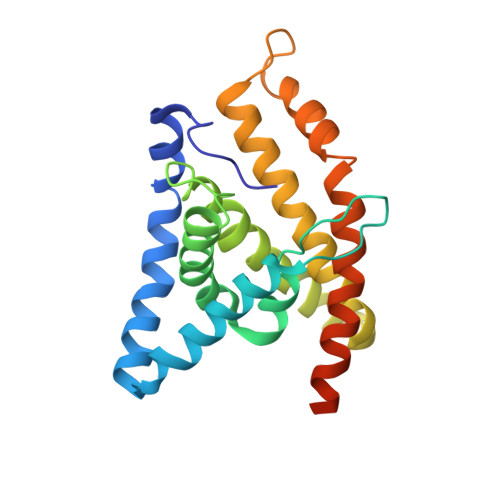
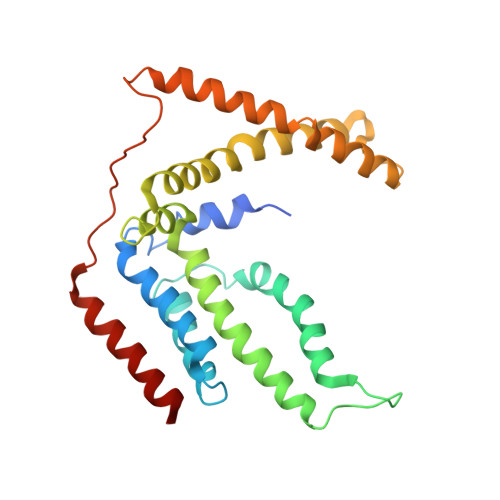
Energy-coupling factor (ECF) transporters are a large family of ATP-binding cassette transporters recently identified in microorganisms. Responsible for micronutrient uptake from the environment, ECF transporters are modular transporters composed of a membrane substrate-binding component EcfS and an ECF module consisting of an integral membrane scaffold component EcfT and two cytoplasmic ATP binding/hydrolysis components EcfA/A'. ECF transporters are classified into groups I and II. Currently, the molecular understanding of group-I ECF transporters is very limited, partly due to a lack of transporter complex structural information. Here, we present structures and structure-based analyses of the group-I cobalt ECF transporter CbiMNQO, whose constituting subunits CbiM/CbiN, CbiQ, and CbiO correspond to the EcfS, EcfT, and EcfA components of group-II ECF transporters, respectively. Through reconstitution of different CbiMNQO subunits and determination of related ATPase and transporter activities, the substrate-binding subunit CbiM was found to stimulate CbiQO's basal ATPase activity. The structure of CbiMQO complex was determined in its inward-open conformation and that of CbiO in β, γ-methyleneadenosine 5'-triphosphate-bound closed conformation. Structure-based analyses revealed interactions between different components, substrate-gating function of the L1 loop of CbiM, and conformational changes of CbiO induced by ATP binding and product release within the CbiMNQO transporter complex. These findings enabled us to propose a working model of the CbiMNQO transporter, in which the transport process requires the rotation or toppling of both CbiQ and CbiM, and CbiN might function in coupling conformational changes between CbiQ and CbiM.
Organizational Affiliation:
National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai 200032, China.