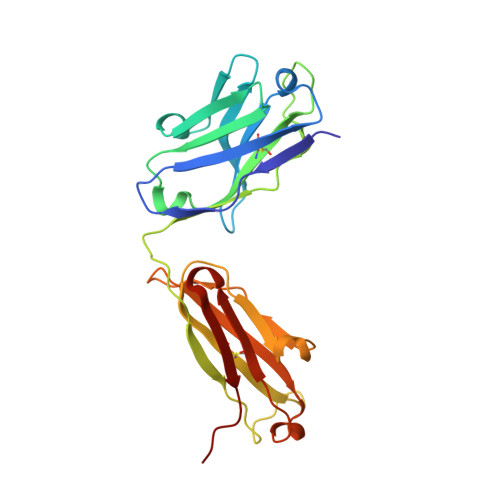
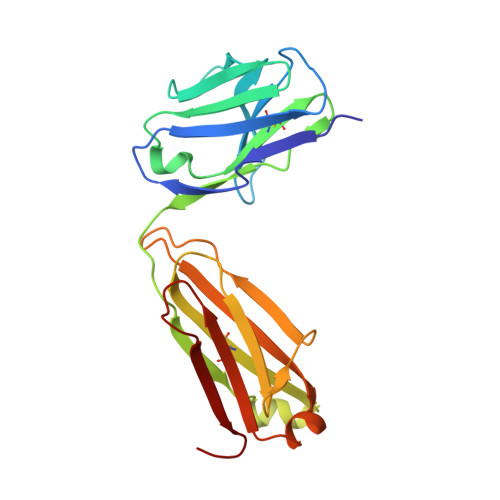
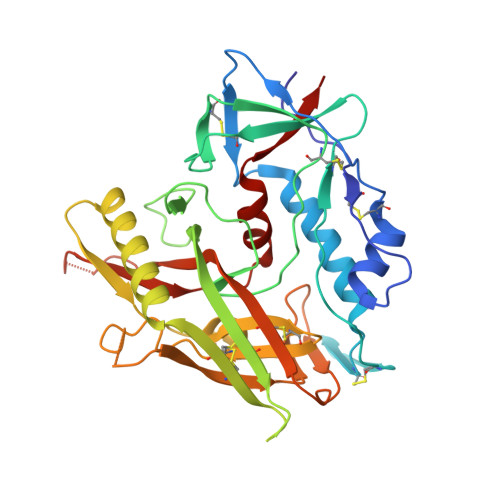
Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth.
Huang, J., Kang, B.H., Ishida, E., Zhou, T., Griesman, T., Sheng, Z., Wu, F., Doria-Rose, N.A., Zhang, B., McKee, K., O'Dell, S., Chuang, G.Y., Druz, A., Georgiev, I.S., Schramm, C.A., Zheng, A., Joyce, M.G., Asokan, M., Ransier, A., Darko, S., Migueles, S.A., Bailer, R.T., Louder, M.K., Alam, S.M., Parks, R., Kelsoe, G., Von Holle, T., Haynes, B.F., Douek, D.C., Hirsch, V., Seaman, M.S., Shapiro, L., Mascola, J.R., Kwong, P.D., Connors, M.(2016) Immunity 45: 1108-1121
- PubMed: 27851912
- DOI: https://doi.org/10.1016/j.immuni.2016.10.027
- Primary Citation of Related Structures:
5TE4, 5TE6, 5TE7 - PubMed Abstract:
Detailed studies of the broadly neutralizing antibodies (bNAbs) that underlie the best available examples of the humoral immune response to HIV are providing important information for the development of therapies and prophylaxis for HIV-1 infection. Here, we report a CD4-binding site (CD4bs) antibody, named N6, that potently neutralized 98% of HIV-1 isolates, including 16 of 20 that were resistant to other members of its class. N6 evolved a mode of recognition such that its binding was not impacted by the loss of individual contacts across the immunoglobulin heavy chain. In addition, structural analysis revealed that the orientation of N6 permitted it to avoid steric clashes with glycans, which is a common mechanism of resistance. Thus, an HIV-1-specific bNAb can achieve potent, near-pan neutralization of HIV-1, making it an attractive candidate for use in therapy and prophylaxis.
Organizational Affiliation:
HIV-Specific Immunity Section of the Laboratory of Immunoregulation, National Institute of Allergy and Infectious Diseases, NIH, Bethesda, MD 20892, USA.