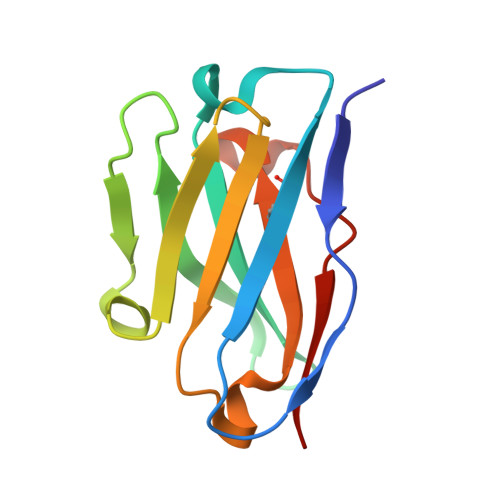
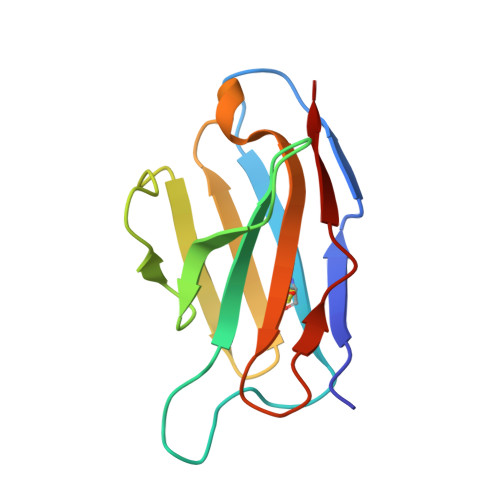
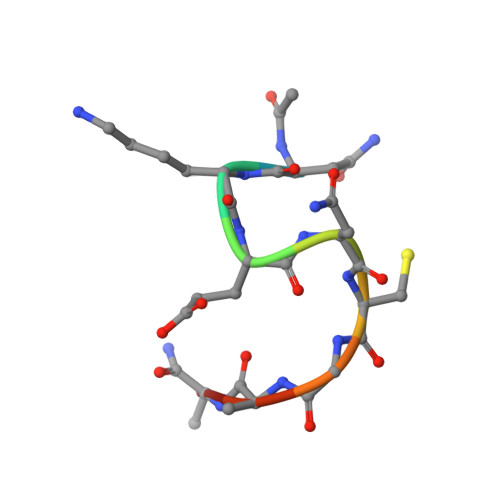
Structure and Characterisation of a Key Epitope in the Conserved C-Terminal Domain of the Malaria Vaccine Candidate MSP2.
Seow, J., Morales, R.A., MacRaild, C.A., Krishnarjuna, B., McGowan, S., Dingjan, T., Jaipuria, G., Rouet, R., Wilde, K.L., Atreya, H.S., Richards, J.S., Anders, R.F., Christ, D., Drinkwater, N., Norton, R.S.(2017) J Mol Biol 429: 836-846
- PubMed: 28189425
- DOI: https://doi.org/10.1016/j.jmb.2017.02.003
- Primary Citation of Related Structures:
5TBD - PubMed Abstract:
Merozoite surface protein 2 (MSP2) is an intrinsically disordered antigen that is abundant on the surface of the malaria parasite Plasmodium falciparum. The two allelic families of MSP2, 3D7 and FC27, differ in their central variable regions, which are flanked by highly conserved C-terminal and N-terminal regions. In a vaccine trial, full-length 3D7 MSP2 induced a strain-specific protective immune response despite the detectable presence of conserved region antibodies. This work focuses on the conserved C-terminal region of MSP2, which includes the only disulphide bond in the protein and encompasses key epitopes recognised by the mouse monoclonal antibodies 4D11 and 9H4. Although the 4D11 and 9H4 epitopes are overlapping, immunofluorescence assays have shown that the mouse monoclonal antibody 4D11 binds to MSP2 on the merozoite surface with a much stronger signal than 9H4. Understanding the structural basis for this antigenic difference between these antibodies will help direct the design of a broad-spectrum and MSP2-based malaria vaccine. 4D11 and 9H4 were reengineered into antibody fragments [variable region fragment (Fv) and single-chain Fv (scFv)] and were validated as suitable models for their full-sized IgG counterparts by surface plasmon resonance and isothermal titration calorimetry. An alanine scan of the 13-residue epitope 3D7-MSP2 207-222 identified the minimal binding epitope of 4D11 and the key residues involved in binding. A 2.2-Å crystal structure of 4D11 Fv bound to the eight-residue epitope NKENCGAA provided valuable insight into the possible conformation of the C-terminal region of MSP2 on the parasite. This work underpins continued efforts to optimise recombinant MSP2 constructs for evaluation as potential vaccine candidates.
Organizational Affiliation:
Monash Institute of Pharmaceutical Sciences, Monash University, Parkville 3052, Australia.