Structural and functional characterization of the hydrogenase-maturation HydF protein.
Caserta, G., Pecqueur, L., Adamska-Venkatesh, A., Papini, C., Roy, S., Artero, V., Atta, M., Reijerse, E., Lubitz, W., Fontecave, M.(2017) Nat Chem Biol 13: 779-784
- PubMed: 28553946
- DOI: https://doi.org/10.1038/nchembio.2385
- Primary Citation of Related Structures:
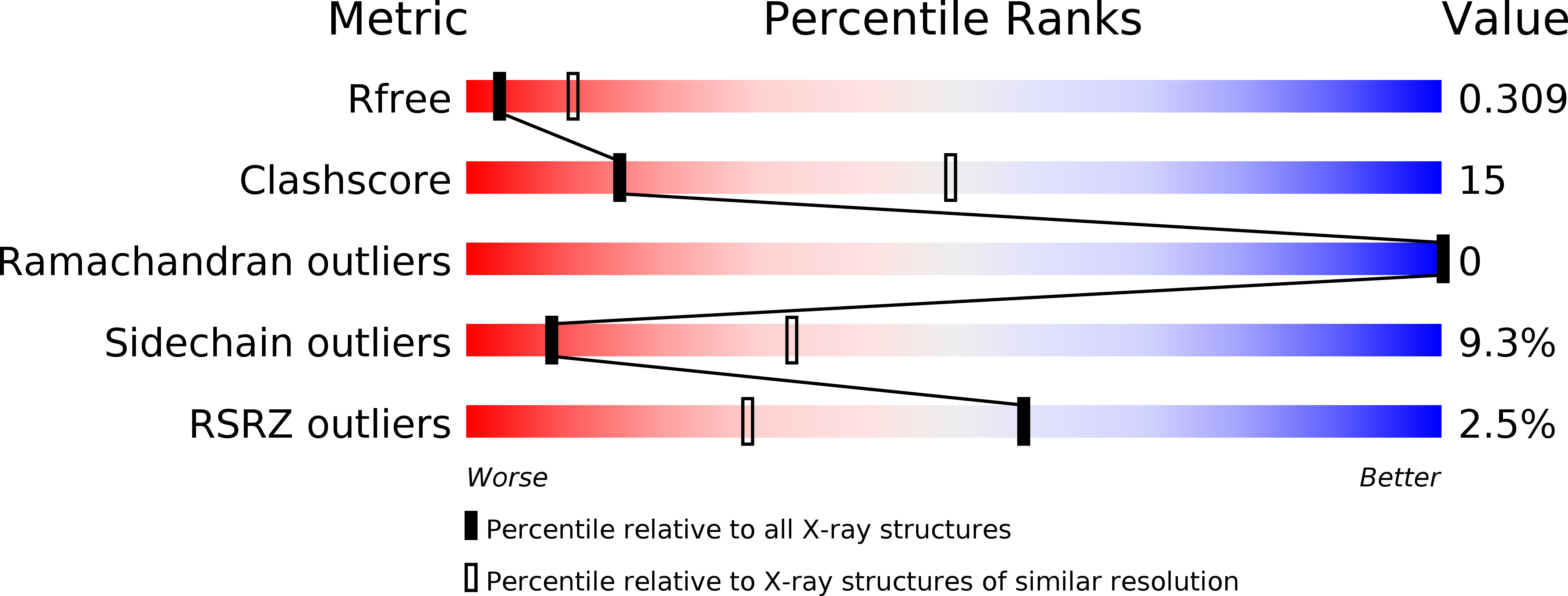
5KH0, 5LAD - PubMed Abstract:
[FeFe] hydrogenase (HydA) catalyzes interconversion between 2H + and H 2 at an active site composed of a [4Fe-4S] cluster linked to a 2Fe subcluster that harbors CO, CN - and azapropanedithiolate (adt 2- ) ligands. HydE, HydG and HydF are the maturases specifically involved in the biosynthesis of the 2Fe subcluster. Using ligands synthesized by HydE and HydG, HydF assembles a di-iron precursor of the 2Fe subcluster and transfers it to HydA for maturation. Here we report the first X-ray structure of HydF with its [4Fe-4S] cluster. The cluster is chelated by three cysteines and an exchangeable glutamate, which allows the binding of synthetic mimics of the 2Fe subcluster. [Fe 2 (adt)(CO) 4 (CN) 2 ] 2- is proposed to be the true di-iron precursor because, when bound to HydF, it matures HydA and displays features in Fourier transform infrared (FTIR) spectra that are similar to those of the native HydF active intermediate. A new route toward the generation of artificial hydrogenases, as combinations of HydF and such biomimetic complexes, is proposed on the basis of the observed hydrogenase activity of chemically modified HydF.
Organizational Affiliation:
Laboratoire de Chimie des Processus Biologiques, Collège de France, Université Pierre et Marie Curie, CNRS UMR 8229, Paris, France.