Crystal structure of an uncharacterized protein (EUBREC_2869) from Eubacterium rectale ATCC 33656 at 1.45 A resolution
Joint Center for Structural Genomics (JCSG)To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Uncharacterized protein | 358 | Agathobacter rectalis ATCC 33656 | Mutation(s): 0 Gene Names: EUBREC_2869 | 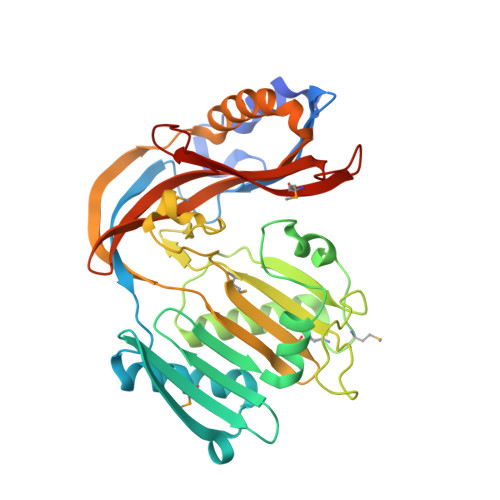 | |
UniProt | |||||
Find proteins for C4ZHW1 (Agathobacter rectalis (strain ATCC 33656 / DSM 3377 / JCM 17463 / KCTC 5835 / VPI 0990)) Explore C4ZHW1 Go to UniProtKB: C4ZHW1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | C4ZHW1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NA Query on NA | B [auth A] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| MSE Query on MSE | A | L-PEPTIDE LINKING | C5 H11 N O2 Se |  | MET |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 47.09 | α = 90 |
| b = 60.06 | β = 90 |
| c = 130.48 | γ = 90 |
| Software Name | Purpose |
|---|---|
| MOSFLM | data reduction |
| SCALA | data scaling |
| SOLVE | phasing |
| BUSTER-TNT | refinement |
| PDB_EXTRACT | data extraction |
| BUSTER | refinement |