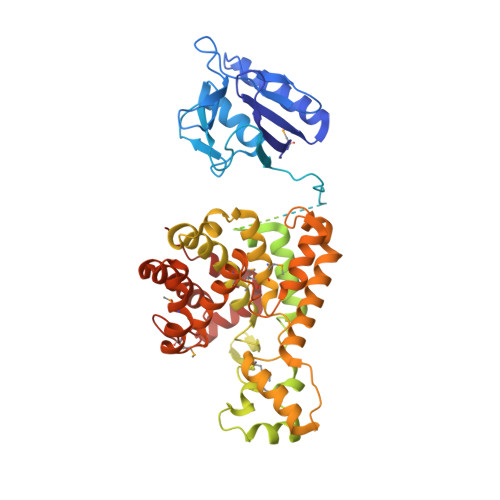
Crystal structure of the Legionella pneumophila lem10 effector reveals a new member of the HD protein superfamily.
Morar, M., Evdokimova, E., Chang, C., Ensminger, A.W., Savchenko, A.(2015) Proteins 83: 2319-2325
- PubMed: 26426142
- DOI: https://doi.org/10.1002/prot.24933
- Primary Citation of Related Structures:
5BQ9 - PubMed Abstract:
Legionella pneumophila, the intracellular pathogen that can cause severe pneumonia known as Legionnaire's disease, translocates close to 300 effectors inside the host cell using Dot/Icm type IVB secretion system. The structure and function for the majority of these effector proteins remains unknown. Here, we present the crystal structure of the L. pneumophila effector Lem10. The structure reveals a multidomain organization with the largest C-terminal domain showing strong structural similarity to the HD protein superfamily representatives. However, Lem10 lacks the catalytic His-Asp residue pair and does not show any in vitro phosphohydrolase enzymatic activity, typical for HD proteins. While the biological function of Lem10 remains elusive, our analysis shows that similar distinct features are shared by a significant number of HD domains found in Legionella proteins, including the SidE family of effectors known to play an important role during infection. Taken together our data point to the presence of a specific group of non-catalytic Legionella HD domains, dubbed LHDs, which are involved in pathogenesis.
Organizational Affiliation:
Department of Chemical Engineering and Applied Chemistry, University of Toronto, Ontario, Canada.