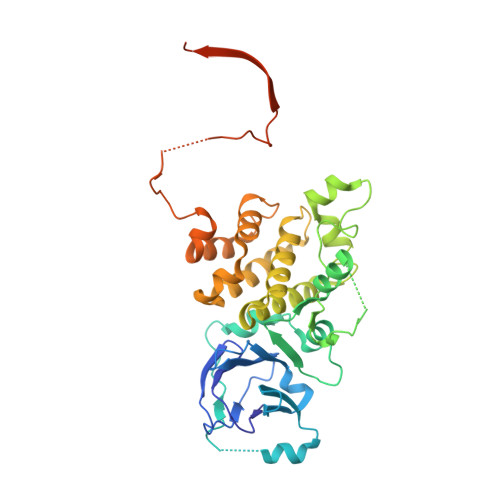
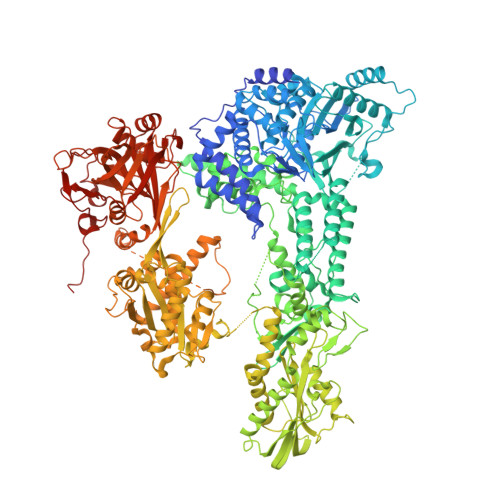
Crystal structure of Prp8 reveals active site cavity of the spliceosome.
Galej, W.P., Oubridge, C., Newman, A.J., Nagai, K.(2013) Nature 493: 638-643
- PubMed: 23354046
- DOI: https://doi.org/10.1038/nature11843
- Primary Citation of Related Structures:
3ZEF, 4I43 - PubMed Abstract:
The active centre of the spliceosome consists of an intricate network formed by U5, U2 and U6 small nuclear RNAs, and a pre-messenger-RNA substrate. Prp8, a component of the U5 small nuclear ribonucleoprotein particle, crosslinks extensively with this RNA catalytic core. Here we present the crystal structure of yeast Prp8 (residues 885-2413) in complex with Aar2, a U5 small nuclear ribonucleoprotein particle assembly factor. The structure reveals tightly associated domains of Prp8 resembling a bacterial group II intron reverse transcriptase and a type II restriction endonuclease. Suppressors of splice-site mutations, and an intron branch-point crosslink, map to a large cavity formed by the reverse transcriptase thumb, and the endonuclease-like and RNaseH-like domains. This cavity is large enough to accommodate the catalytic core of group II intron RNA. The structure provides crucial insights into the architecture of the spliceosome active site, and reinforces the notion that nuclear pre-mRNA splicing and group II intron splicing have a common origin.
Organizational Affiliation:
MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge CB2 0QH, UK.