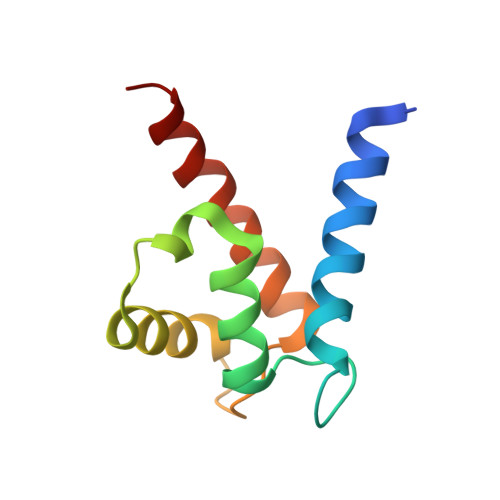
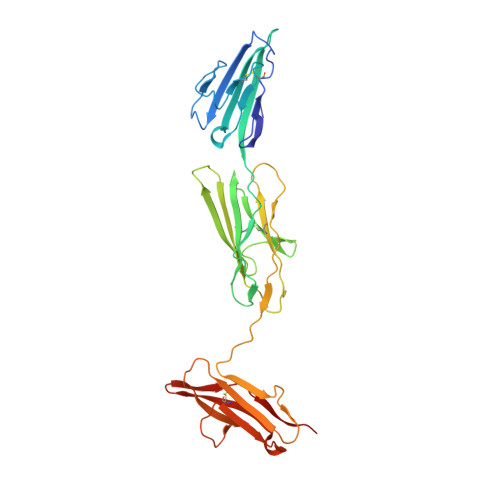The Structure of the RAGE:S100A6 Complex Reveals a Unique Mode of Homodimerization for S100 Proteins.
Yatime, L., Betzer, C., Jensen, R.K., Mortensen, S., Jensen, P.H., Andersen, G.R.(2016) Structure 24: 2043-2052
- PubMed: 27818100
- DOI: https://doi.org/10.1016/j.str.2016.09.011
- Primary Citation of Related Structures:
4P2Y, 4YBH - PubMed Abstract:
S100 proteins are calcium-dependent regulators of homeostatic processes. Upon cellular response to stress, and notably during tumorigenesis, they relocalize to the extracellular environment where they induce pro-inflammatory signals by activating the receptor for advanced glycation end products (RAGE), thereby facilitating tumor growth and metastasis. Despite its importance in sustaining inflammation, the structural basis for RAGE-S100 crosstalk is still unknown. Here we report two crystal structures of the RAGE:S100A6 complex encompassing a full-length RAGE ectodomain. The structures, in combination with a comprehensive interaction analysis, suggest that the primary S100A6 binding site is formed by the RAGE C1 domain. Complex formation with S100A6 induces a unique dimeric conformation of RAGE that appears suited for signal transduction and intracellular effector recruitment. Intriguingly, S100A6 adopts a dimeric conformation radically different from all known S100 dimers. We discuss the physiological relevance of this non-canonical homodimeric form in vivo.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10C, 8000 Aarhus, Denmark. Electronic address: laure.yatime@inserm.fr.