Crystal structure of cindoxin, the P450cin redox partner.
Madrona, Y., Hollingsworth, S.A., Tripathi, S., Fields, J.B., Rwigema, J.C., Tobias, D.J., Poulos, T.L.(2014) Biochemistry 53: 1435-1446
- PubMed: 24533927
- DOI: https://doi.org/10.1021/bi500010m
- Primary Citation of Related Structures:
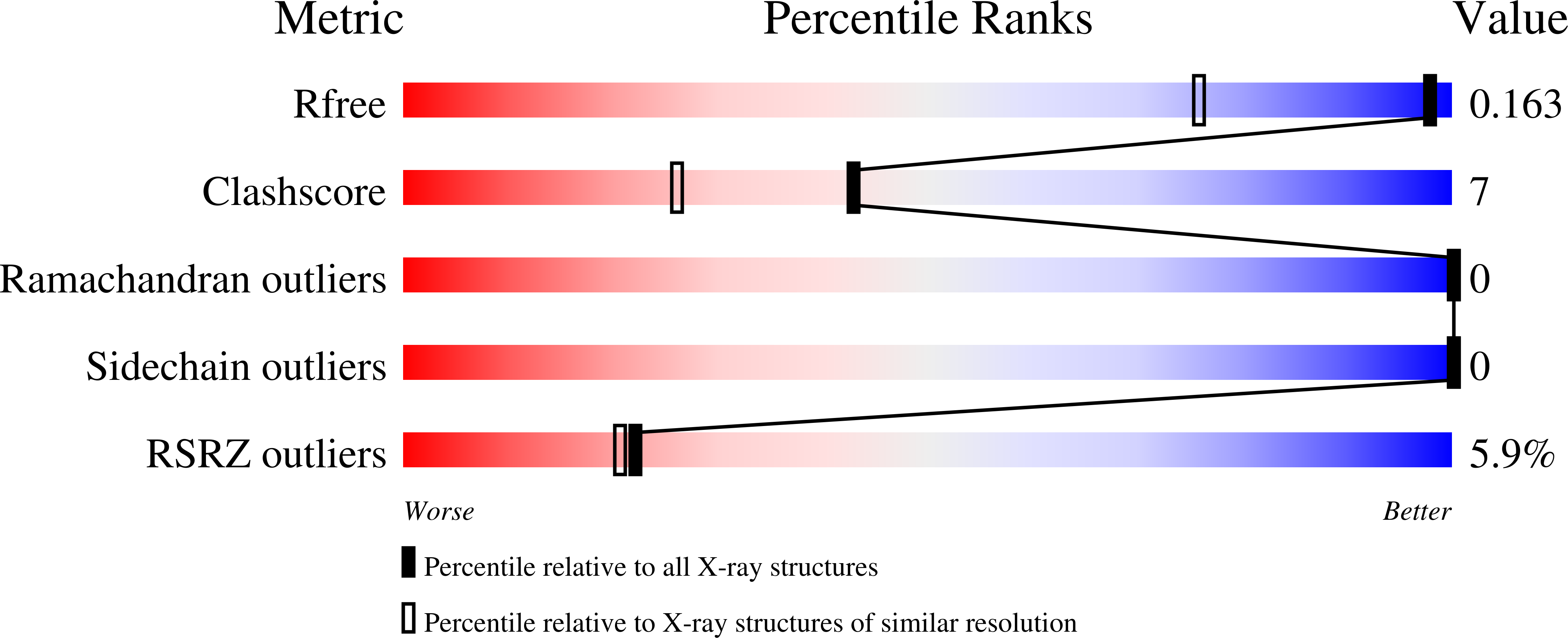
4OXX - PubMed Abstract:
The crystal structure of the flavin mononucleotide (FMN)-containing redox partner to P450cin, cindoxin (Cdx), has been determined to 1.3 Å resolution. The overall structure is similar to that of the FMN domain of human cytochrome P450 reductase. A Brownian dynamics-molecular dynamics docking method was used to produce a model of Cdx with its redox partner, P450cin. This Cdx-P450cin model highlights the potential importance of Cdx Tyr96 in bridging the FMN and heme cofactors as well P450cin Arg102 and Arg346. Each of the single-site Ala mutants exhibits ~10% of the wild-type activity, thus demonstrating the importance of these residues for binding and/or electron transfer. In the well-studied P450cam system, redox partner binding stabilizes the open low-spin conformation of P450cam and greatly decreases the stability of the oxy complex. In sharp contrast, Cdx does not shift P450cin to a low-spin state, although the stability of oxy-P450cin is decreased 10-fold in the presence of Cdx. This indicates that Cdx may have a modest effect on the open-closed equilibrium in P450cin compared to that in P450cam. It has been postulated that part of the effector role of Pdx on P450cam is to promote a significant structural change that makes available a proton relay network involving Asp251 required for O2 activation. The structure around the corresponding Asp in P450cin, Asp241, provides a possible structural reason for why P450cin is less dependent on its redox partner for functionally important structural changes.
Organizational Affiliation:
Departments of †Molecular Biology and Biochemistry, ‡Chemistry, and §Pharmaceutical Sciences, University of California , Irvine, California 92697-3900, United States.