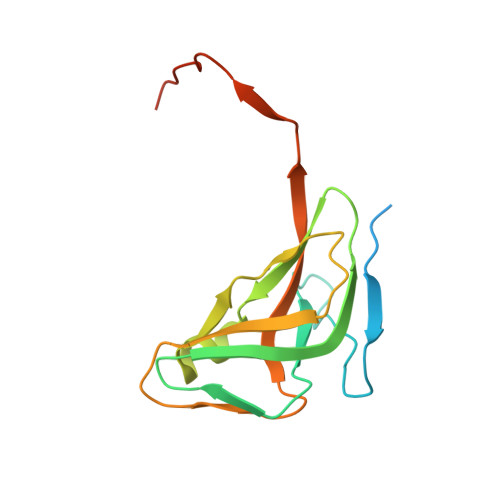
Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation.
Inoguchi, N., Chaiseeda, K., Yamanishi, M., Kim, M.K., Jang, Y., Bajaj, M., Chia, C.P., Becker, D.F., Moriyama, H.(2015) BMC Res Notes 8: 784-784
- PubMed: 26666293
- DOI: https://doi.org/10.1186/s13104-015-1760-1
- Primary Citation of Related Structures:
4OOP, 4OOQ - PubMed Abstract:
Deoxyuridine triphosphate nucleotidohydrolase (dUTPase) hydrolyzes dUTP to dUMP and pyrophosphate to maintain the cellular thymine-uracil ratio. dUTPase is also a target for cancer chemotherapy. However, the mechanism defining its substrate affinity remains unclear. Sequence comparisons of various dUTPases revealed that Arabidopsis thaliana dUTPase has a unique tryptophan at position 93, which potentially contributes to its degree of substrate affinity. To better understand the roles of tryptophan 93, A. thaliana dUTPase was studied.
Organizational Affiliation:
School of Biological Sciences, University of Nebraska-Lincoln, Lincoln, NE, USA. ninoguchi@huskers.unl.edu.