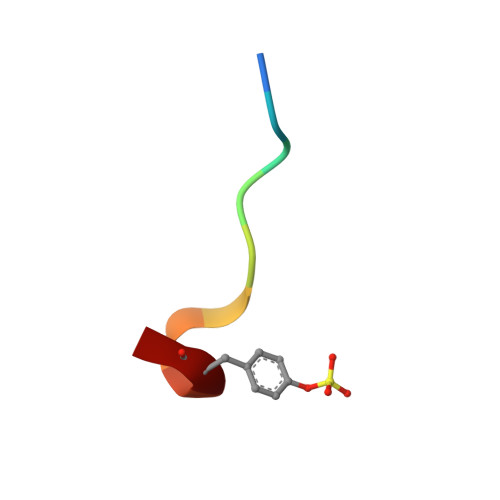
Discovery of benzothiazole guanidines as novel inhibitors of thrombin and trypsin IV.
Karle, M., Knecht, W., Xue, Y.(2012) Bioorg Med Chem Lett 22: 4839-4843
- PubMed: 22726924
- DOI: https://doi.org/10.1016/j.bmcl.2012.05.046
- Primary Citation of Related Structures:
3PO1 - PubMed Abstract:
In a project to find novel neutral P1 fragments for the synthesis of thrombin inhibitors with improved pharmacokinetic properties, fragments containing a benzothiazole guanidine scaffold were identified as weak thrombin inhibitors. WaterLOGSY (Water-Ligand Observed via Gradient SpectroscopY) NMR was used to detect fragments binding to thrombin and these fragments were followed up by Biacore A100 affinity measurements and enzyme assays. A crystal structure of the most potent compound with thrombin was obtained and revealed an unexpected binding mode as well as the key interactions of the fragment with the protein. Based on these results, the structure-based design and synthesis of a small series of optimized novel substituted benzothiazole guanidines with comparatively low pK(a) values was accomplished. Testing of these compounds against human trypsin I and human trypsin IV revealed unexpected inhibitory activity and selectivity of some of the compounds, making them attractive starting points for selective trypsin inhibitors.
Organizational Affiliation:
AstraZeneca R&D Mölndal, SE-43183 Mölndal, Sweden. Michael.Karle@astrazeneca.com