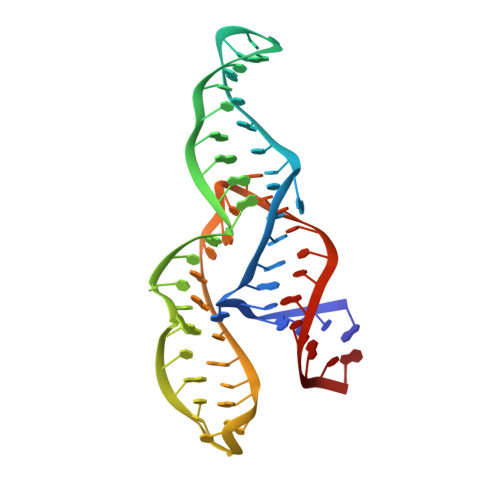
The NMR structure of the II-III-VI three-way junction from the Neurospora VS ribozyme reveals a critical tertiary interaction and provides new insights into the global ribozyme structure.
Bonneau, E., Girard, N., Lemieux, S., Legault, P.(2015) RNA 21: 1621-1632
- PubMed: 26124200
- DOI: https://doi.org/10.1261/rna.052076.115
- Primary Citation of Related Structures:
2N3Q, 2N3R - PubMed Abstract:
As part of an effort to structurally characterize the complete Neurospora VS ribozyme, NMR solution structures of several subdomains have been previously determined, including the internal loops of domains I and VI, the I/V kissing-loop interaction and the III-IV-V junction. Here, we expand this work by determining the NMR structure of a 62-nucleotide RNA (J236) that encompasses the VS ribozyme II-III-VI three-way junction and its adjoining stems. In addition, we localize Mg(2+)-binding sites within this structure using Mn(2+)-induced paramagnetic relaxation enhancement. The NMR structure of the J236 RNA displays a family C topology with a compact core stabilized by continuous stacking of stems II and III, a cis WC/WC G•A base pair, two base triples and two Mg(2+) ions. Moreover, it reveals a remote tertiary interaction between the adenine bulges of stems II and VI. Additional NMR studies demonstrate that both this bulge-bulge interaction and Mg(2+) ions are critical for the stable folding of the II-III-VI junction. The NMR structure of the J236 RNA is consistent with biochemical studies on the complete VS ribozyme, but not with biophysical studies performed with a minimal II-III-VI junction that does not contain the II-VI bulge-bulge interaction. Together with previous NMR studies, our findings provide important new insights into the three-dimensional architecture of this unique ribozyme.
Organizational Affiliation:
Département de Biochimie et Médecine Moléculaire, Université de Montréal, Montréal, Quebec H3C 3J7, Canada.