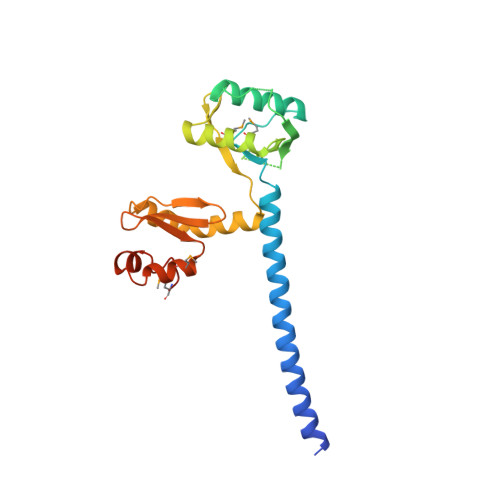
Trimeric Structure and Flexibility of the L1Orf1P Protein in Human L1 Retrotransposition
Khazina, E., Truffault, V., Buettner, R., Schmidt, S., Coles, M., Weichenrieder, O.(2011) Nat Struct Mol Biol 18: 1006
- PubMed: 21822284
- DOI: https://doi.org/10.1038/nsmb.2097
- Primary Citation of Related Structures:
2LDY, 2YKO, 2YKP, 2YKQ - PubMed Abstract:
The LINE-1 (L1) retrotransposon emerges as a major source of human interindividual genetic variation, with important implications for evolution and disease. L1 retrotransposition is poorly understood at the molecular level, and the mechanistic details and evolutionary origin of the L1-encoded L1ORF1 protein (L1ORF1p) are particularly obscure. Here three crystal structures of trimeric L1ORF1p and NMR solution structures of individual domains reveal a sophisticated and highly structured, yet remarkably flexible, RNA-packaging protein. It trimerizes via an N-terminal, ion-containing coiled coil that serves as scaffold for the flexible attachment of the central RRM and the C-terminal CTD domains. The structures explain the specificity for single-stranded RNA substrates, and a mutational analysis indicates that the precise control of domain flexibility is critical for retrotransposition. Although the evolutionary origin of L1ORF1p remains unclear, our data reveal previously undetected structural and functional parallels to viral proteins.
Organizational Affiliation:
Department of Biochemistry, Max Planck Institute for Developmental Biology, Tübingen, Germany.