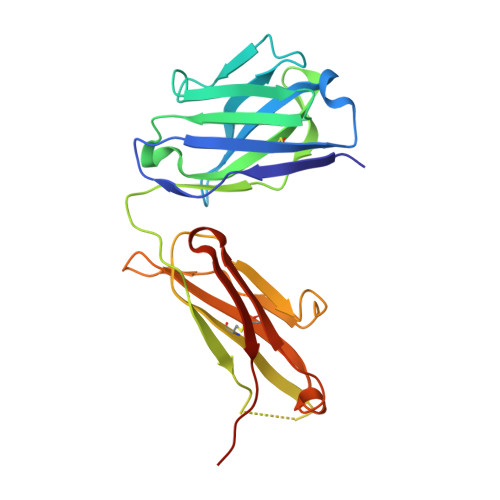
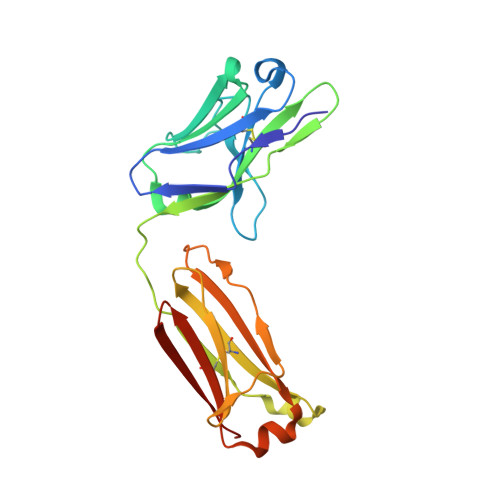
Structure of Il-17A in Complex with a Potent, Fully Human Neutralising Antibody.
Gerhardt, S., Abbott, W.M., Hargreaves, D., Pauptit, R.A., Davies, R.A., Needham, M.R., Langham, C., Barker, W., Aziz, A., Snow, M.J., Dawson, S., Welsh, F., Wilkinson, T., Vaugan, T., Beste, G., Bishop, S., Popovic, B., Rees, G., Sleeman, M., Tuske, S.J., Coales, S.J., Hamuro, Y., Russell, C.(2009) J Mol Biol 394: 905
- PubMed: 19835883
- DOI: https://doi.org/10.1016/j.jmb.2009.10.008
- Primary Citation of Related Structures:
2VXS - PubMed Abstract:
IL-17A is a pro-inflammatory cytokine produced by the newly identified Th17 subset of T-cells. We have isolated a human monoclonal antibody to IL-17A (CAT-2200) that can potently neutralize the effects of recombinant and native human IL-17A. We determined the crystal structure of IL-17A in complex with the CAT-2200 Fab at 2.6 A resolution in order to provide a definitive characterization of the epitope and paratope regions. Approximately a third of the IL-17A dimer is disordered in this crystal structure. The disorder occurs in both independent copies of the complex in the asymmetric unit and does not appear to be influenced by crystal packing. The complex contains one IL-17A dimer sandwiched between two CAT-2200 Fab fragments. The IL-17A is a disulfide-linked homodimer that is similar in structure to IL-17F, adopting a cystine-knot fold. The structure is not inconsistent with the previous prediction of a receptor binding cavity on IL-17 family members. The epitope recognized by CAT-2200 is shown to involve 12 amino acid residues from the quaternary structure of IL-17A, with each Fab contacting both monomers in the dimer. All complementarity-determining regions (CDRs) in the Fab contribute to a total of 16 amino acid residues in the antibody paratope. In vitro affinity optimization was used to generate CAT-2200 from a parental lead antibody using random mutagenesis of CDR3 loops. This resulted in seven amino acid changes (three in VL-CDR3 and four in VH-CDR3) and gave an approximate 30-fold increase in potency in a cell-based neutralization assay. Two of the seven amino acids form part of the CAT-2200 paratope. The observed interaction site between CAT-2200 and IL-17A is consistent with data from hydrogen/deuterium exchange mass spectrometry and mutagenesis approaches.
Organizational Affiliation:
AstraZeneca, Alderley Park, Macclesfield, Cheshire SK10 4TG, UK.