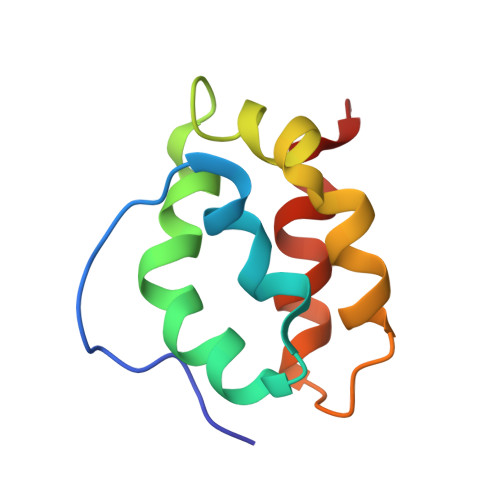
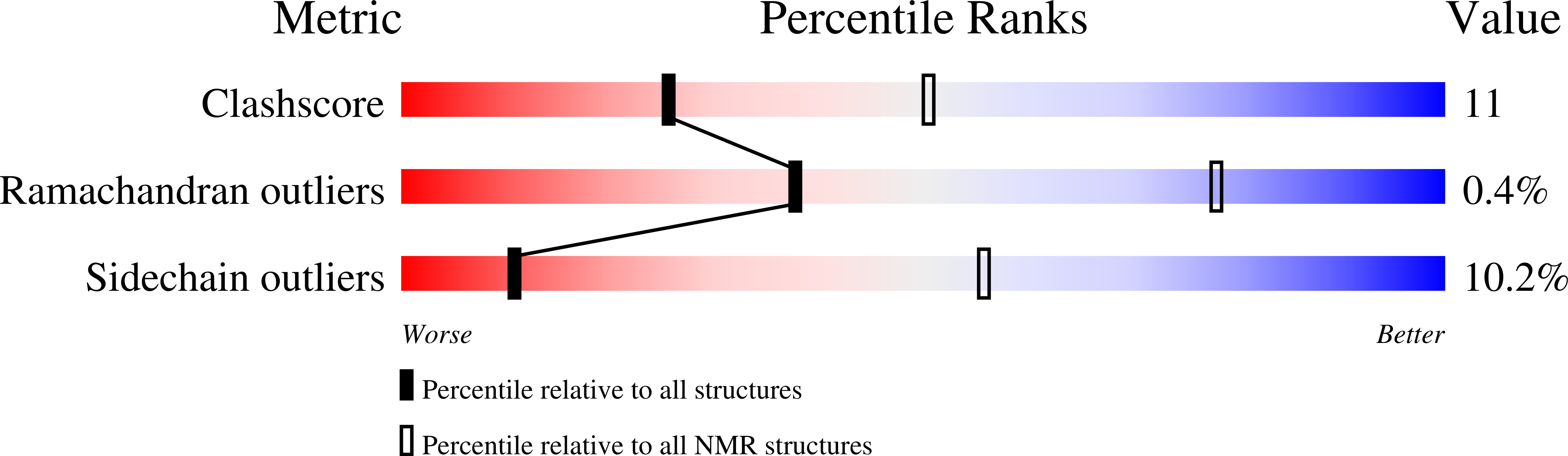Spontaneous self-assembly of engineered armadillo repeat protein fragments into a folded structure
Watson, R.P., Christen, M.T., Ewald, C., Bumbak, F., Reichen, C., Mihajlovic, M., Schmidt, E., Guntert, P., Caflisch, A., Pluckthun, A., Zerbe, O.(2014) Structure 22: 985-995
- PubMed: 24931467
- DOI: https://doi.org/10.1016/j.str.2014.05.002
- Primary Citation of Related Structures:
2RU4, 2RU5 - PubMed Abstract:
Repeat proteins are built of modules, each of which constitutes a structural motif. We have investigated whether fragments of a designed consensus armadillo repeat protein (ArmRP) recognize each other. We examined a split ArmRP consisting of an N-capping repeat (denoted Y), three internal repeats (M), and a C-capping repeat (A). We demonstrate that the C-terminal MA fragment adopts a fold similar to the corresponding part of the entire protein. In contrast, the N-terminal YM2 fragment constitutes a molten globule. The two fragments form a 1:1 YM2:MA complex with a nanomolar dissociation constant essentially identical to the crystal structure of the continuous YM3A protein. Molecular dynamics simulations show that the complex is structurally stable over a 1 μs timescale and reveal the importance of hydrophobic contacts across the interface. We propose that the existence of a stable complex recapitulates possible intermediates in the early evolution of these repeat proteins.
Organizational Affiliation:
Department of Chemistry, University of Zürich, 8057 Zürich, Switzerland.